
Group photo.From left to right. Francisco García del Portillo, M. Graciela Pucciarelli, Pablo García, Estel Ramos, Diana Barroso, Gadea Rico, Daniela Gargano and Noelia López.
Many diseases that impact the health of man and animals are caused by bacteria that invade and colonize the intracellular niche of eukaryotic cells. These diseases include, among others, tuberculosis, typhoid fever, listeriosis, chlamydiosis, brucellosis, rickettiosis, Q fever and legionellosis. Some pathogenic bacteria that produce these pathologies, such as those of the genera Rickettsia ., 2016), so they can be prescribed as an alternative or complementary treatment of infections by Chamydia spp. y Coxiella spp., are unable to grow and proliferate outside the eukaryotic cell.
Of great research interest are also the ‘facultative’ intracellular pathogenic bacteria that colonize different eukaryotic cell types as well as diverse environments outside the host. The data obtained to date show the existence of regulation systems that are responsible for reprogramming the physiology of the bacteria during the transition from the extracellular to the intracellular environment, or vice versa. The main objective of our group is to understand these regulation patterns and, therefore, the mechanisms of adaptation to the intracellular lifestyle once the pathogen has invaded the eukaryotic cell. To undertake this undertaking, we used two bacteria with different envelopes and a varied intracellular way of life. On the one hand, Gram-positive bacteria Listeria monocytogenes, causative agent of listeriosis. L. monocytogenes It crosses natural defense barriers such as the intestinal epithelium, the blood-brain barrier and the placenta. As a second model pathogen, Gram-negative bacteria Samonella enterica serovar Typhimurium (S. Typhimurium), the causative agent of gastroenteritis and, eventually, of systemic disease if there are additional risks such as coinfection or alterations in host defenses. Both pathogens initiate infection in cattle and humans after consumption of contaminated feed.
The bacterial cell wall and adaptation to the eukaryotic intracellular environment
A key cellular structure in maintaining the shape and integrity of almost all bacteria is the 'wall' or envelope. Within the wall is the peptidoglycan macromolecule, also known as 'murein', which ensures cell shape and integrity. Historically, the biochemistry of peptidoglycan has been intensively studied for being the target of antibiotics such as those of the beta-lactam group. More recent studies indicate that peptidoglycan is an alarm signal for the host's innate defense system. Thus, receptors have been characterized located in the cytosol of eukaryotic cells, such as Nod1 and Nod2, which recognize 'molecular patterns' present in peptidoglycan fragments. Interestingly, this recognition occurs inside the eukaryotic cell, indicating that evolution seems to have designed defenses to control intracellular bacterial infections. Of interest, certain intracellular bacterial pathogens have evolved by making modifications in the peptidoglycan that influence its recognition by defense enzymes (eg lysozyme) or Nod-type receptors. In the context of our research, we are also interested in a large family of proteins covalently linked to peptidoglycan in the genus Listeria. The vast majority of these surface proteins (they appear in an average of 40 in all species and lineages of this genus with sequenced genome) are exclusive to Listeria, existing an appreciable number of the same ones that are present only in pathogenic species. The function of approximately 90% of these proteins is unknown, although it is postulated that they could be involved in the adaptation of Listeria to diverse environments.
Our research in L. monocytogenes and S. enterica serovar Typhimurium
The experimental approaches used by our group have a common denominator, the "Isolation of bacteria from infected eukaryotic cells in culture". This methodology is laborious in terms of the number of eukaryotic cells that must be infected to obtain the amount of bacterial material necessary to perform molecular biology. As an illustrative example, obtaining 10Ferrándiz MJ, Martín-Galiano AJ, Arnanz C, Camacho-Soguero I, Tirado-Vélez JM, de la Campa AG. intracellular bacteria (the equivalent of 10 microliters of a bacterial culture that has grown overnight in laboratory medium!!!) means infecting approximately 10Ferrándiz MJ, Martín-Galiano AJ, Arnanz C, Camacho-Soguero I, Tirado-Vélez JM, de la Campa AG. fibroblasts or epithelial cells. Reaching such a quantity of eukaryotic cells in culture requires at least a week of work. When the experiment is aimed at the purification of peptidoglycan synthesized by intracellular bacteria, these numbers increase by several orders of magnitude. Despite these difficulties, this experimental approach is the only one that allows obtaining information on the biology of the pathogen during its intracellular infection cycle. Thus, we have obtained achievements such as the transcriptome of S. Typhimurium when it persists in a non-growing intracellular state; the structure of peptidoglycan S. Typhimurium synthesizes inside epithelial cells; the wall proteome of L. monocytogenes isolated from infected epithelial cells; and, the expression profile in real time and throughout different phases of intracellular infection of 56 small regulatory RNAs (sRNA) of S. Typhimurium.
The ‘global nature’ information obtained in these studies currently allows us to address questions with a similar methodology that we now address to proteins, regulatory RNAs and specific processes. As an example, we have deciphered a regulation that acts on a protein of L. monocytogenes bound to peptidoglycan that the pathogen induces in the intracellular environment. Increasing the levels of this protein requires the binding of a regulatory sRNA to a 5'-UTR (untranslated region of mRNA) of a variant of the target gene transcript. On S. Typhimurium we are studying enzymes involved in the synthesis, remodeling and hydrolysis of peptidoglycan in intracellular bacteria, always taking bacteria grown in laboratory media as a reference. Within this group of enzymes we have identified some unique to the genus Salmonella. Interestingly, some of them are positively regulated in the intracellular environment.
Also note our interest in knowing the 'cellular biology' of intracellular infection, focusing on a model of persistent infection in which the bacterium does not proliferate in the eukaryotic cell. The data obtained to date implicate the autophagic machinery of the host cell as a growth regulator of S. Typhimurium. In the case of persistent infection, the autophagy of the intracellular pathogen shows distinctive characteristics from those described in the literature in other models of infection.
Highlight as a final message the extraordinary differences that we observe in many processes when we study these bacteria in the intracellular and extracellular environment. Understanding how, when, and who is responsible for these changes will undoubtedly maintain our enthusiasm for that still-so-mysterious "life" that certain pathogenic bacteria develop inside our cells.
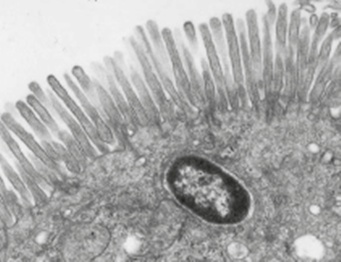
Figure 1. Image of Salmonella enterica serovar Typhimurium after invading an enterocyte of the mouse intestinal epithelium. Note that the bacterium is located in a vacuolar compartment, also known as a phagosome or SCV, for 'Salmonella-containing vacuole'. Unlike this intracellular lifestyle, Listeria monocytogenes uses proteins that degrade the phagosomal membrane, subsequently proliferating in the cytosol of the infected cell.
Representative bibliography
Ortega AD, Quereda JJ, Pucciarelli, MG and García-del Portillo, F. (2014). Non-coding RNA regulation in pathogenic bacteria located inside eukaryotic cells. Front CellInfectMicrobiol. (en prensa)
Quereda JJ, Ortega AD, Pucciarelli, MG and García-del Portillo, F. (2014). The Listeria small RNA Rli27 regulates a cell wall protein inside eukaryotic cells by targeting a long 5′-UTR variant. PLoSGenetics (en prensa)
Hernández, SB, Cava F, Pucciarelli MG, García-del Portillo F., de Pedro MA and Casadesús J. (2014). Bile-induced peptidoglycan remodelling in Salmonella enterica. About. Microbiol. doi: 10.1111 / 1462-2920.12491.
Bécavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kühbacher A, Brisse S, Pucciarelli MG, García-Del Portillo F., Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro -Cerdá J, Moszer I, Bierne H and Cossart P. (2014) Comparison of widely used Listeria monocytogenes strains EGD, 10403S and EGD-e highlights genomic divergence underlying differences in pathogenicity. mBIO doi: 10.1128 / mBio.00969-14.
Mariscotti JF, Quereda JJ, Garcia-Del Portillo F, Pucciarelli MG. (2014) The Listeria monocytogenes LPXTG surface protein Lm01413 is an invasin with capacity to bind mucin. Int. J. Med. Microbiol. 304: 393-404.
Quereda JJ and Pucciarelli MG (2014) Deletion of the membrane protein Lmo0412 increases the virulence of Listeria monocytogenes. MicrobesInfect. 16: 623-632.
Núñez-Hernández C, Alonso A, Pucciarelli MG, Casadesús J and García-Del Portillo F. (2014). Dormant intracellular Salmonella discriminates among SPI-2 effectors to persist inside fibroblasts. Infect. Immun. 82: 221-232.
Gonzalo-Asensio J, Ortega AD, Rico-Pérez G, Pucciarelli MG and García-Del Portillo F (2013). A Novel antisense RNA from the Salmonella virulence plasmid pSLT expressed by non-growing bacteria inside eukaryotic cells. PLoS One 8(10):e77939. doi: 10.1371/journal.pone.0077939.Microbiol. 16: 87-92.
Silva , IJ , Ortega AD , Viegas SC , Garcia-del Portillo F (*) and Arraiano CM. (2013). An RpoS-dependent sRNA regulates the expression of a chaperone involved in protein folding. RNA 19: 1253-1265. (*) co-corresponding author.
Hernández, S.B., Ayala, J.A., Rico-Pérez, G., García-del Portillo, F., Casadesús, J. (2013) Increased bile resistance in Salmonella enterica mutants lacking Prc periplasmic protease Int. Microbiol. 2013 Jun;16(2):87-92.
Núñez-Hernández C, Tierrez A, Ortega AD, Pucciarelli MG, Godoy M, Esiman B, Casadesús J and García-del Portillo, F. (2013). Genome expression analysis of non-proliferating intracellular Salmonella unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy. Infect. Immun.81:154-165.
Ortega, A.D., Gonzalo-Asensio, J. and García-del Portillo, F. (2013). Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA Biology 9: 469-488.
García-del Portillo F, Calvo E, D'Orazio V y Pucciarelli MG (2011). Association of ActA to peptidoglycan revealed by cell wall proteomics of intracelular Listeria monocytogenes. J. Biol. Chem. 286: 34675-34689.
Aiastui A, Pucciarelli MG and García-del Portillo F. (2010). Salmonella invades fibroblasts by multiple routes differing from the entry into epithelial cells. Infect. Immun. 78: 2700-2713.
