
Group photo. From left to right. Luis M. Mateos, Alfonso Gonzalo, Miriam Arienza, Almudena F. Villadangos, José A. Gil.
The research group led by doctors José A. Gil and Luis M. Mateos, called "Molecular Biology of Corinebacteria" (http://microbio.unileon.es/wordpress), studies various aspects of the Molecular Biology of corynebacteria Corynebacterium glutamicum, an actinobacteria that owes the species epithet "glutamicum”To its ability to produce glutamic acid. During the last fifteen years, the group has focused its research on: A) the study of cell division processes in C. glutamicum, in order to use this knowledge for the design of antimicrobial agents that control diseases caused by corynebacteria and pathogenic mycobacteria; B) analysis of arsenic resistance in C. glutamicum with the aim of knowing the metal detoxifying systems and their effect on cell stress, and the achievement of microorganisms capable of being used as biocontainers of heavy metals and applied in bioremediation.
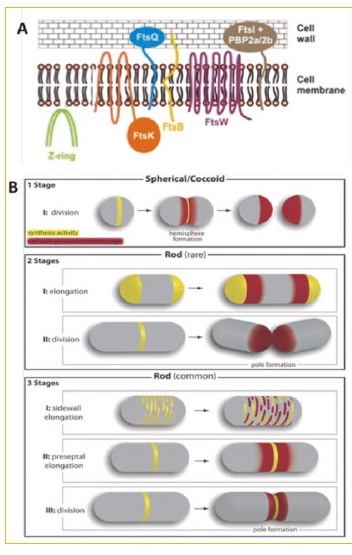 Figure 1. A) Minimalist division machinery of Corynebacterium glutamicum. B) Elongation / division models in different bacteria. (1 stage). Growth at the level of the dividing septum in coccoid bacteria lacking MreB such as Streptococcus pneumoniae (2 stages). Polar growth in bacillary bacteria lacking MreB such as C. glutamicom or Mycobacterium tuberculosis. (3 stages) MreB-driven growth along the lateral wall of B. subtilis or E. coli. The yellow color shows the sites of new peptidoglycan synthesis and the red color shows the site where the new peptidoglycan is inserted. (http://jcb.rupress.org/content/179/3/381.full)
Figure 1. A) Minimalist division machinery of Corynebacterium glutamicum. B) Elongation / division models in different bacteria. (1 stage). Growth at the level of the dividing septum in coccoid bacteria lacking MreB such as Streptococcus pneumoniae (2 stages). Polar growth in bacillary bacteria lacking MreB such as C. glutamicom or Mycobacterium tuberculosis. (3 stages) MreB-driven growth along the lateral wall of B. subtilis or E. coli. The yellow color shows the sites of new peptidoglycan synthesis and the red color shows the site where the new peptidoglycan is inserted. (http://jcb.rupress.org/content/179/3/381.full)Cellular division
We have studied the cell division / elongation process of C. glutamicum since it has a “minimalist” division machinery when compared to the machinery of Escherichia coli (Fig. 1.A), that is, it lacks many of the proteins (FtsA, FtsL, FtsN, AmiC, MinCDE, ZipA, MreB ...) that are considered essential in the cell division machinery of E. coli. During these last years we have studied the role of the DivIVA protein in the cell elongation process (Letek et al., 2008; Valbuena et al., 2007), as well as the role of serine-threonine-protein kinases in this process.
(Fiuza et al., 2008a and 2008b). We also describe the presence of an intermediate filament (RsmP) in C. glutamicum regulated by phosphorylation and involved in the maintenance of bacillary morphology (Fiuza et al., 2010) glutamicum shows a "rare" growth model that differentiates them from the classic models of E. coli, Bacillus subtilis o Streptococcus pneumoniae (Fig. 1.B). The results obtained with C. glutamicum will allow us to design compounds that inhibit cell division in related pathogenic organisms such as C. diphtheriae O M. tuberculosis.
Bacterial resistance to arsenic
The other line of research has been based on the arsenic-bacteria interaction; Arsenic has been present in the atmosphere since the formation of the Earth and living beings have been continuously subjected to this agent, which is why the presence of detoxification systems or resistance to arsenic is quite frequent (Mateos et al., 2006). C. glutamicum is no exception and has arsenic resistance genes that are grouped into two operons ars, each of them encoding for a regulator / repressor (ArsR), an arsenate reductase protein (ArsC) and an arsenite permease (Acr3).
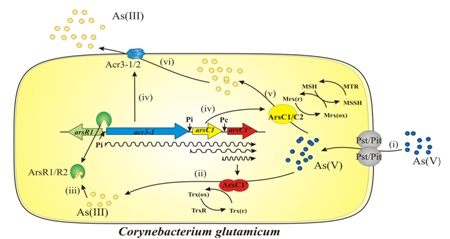
Figure 2. Arsenic resistance model in C. glutanicum. (i) Arsenate input [As (V)]; (ii) constitutive expression of the ArsC1 protein and reduction of As (V) to arsenite [As (III)]; (iii) binding of As (III) to regulators (ArsR1 / R2) and derepression of the ars operon; (iv) transcription of the genes encoding for arsenate reductases (ArsC1 / C2 / C1 ′); (v) reduction of As (V) by ArsC1 and ArsC2 reductases using the MSH / Mrx1 system and (vi) release of As (III) formed in the cell through the arsenite permeases Acr3-1 / 2. (Viladangos et al., 2011).
From the analysis of the genes and the corresponding detoxifying proteins, mutant and recombinant clones of C. glutamicum capable of accumulating up to 30 times more arsenic than the wild strain and that could be used in bioremediation processes (Ordóñez et al., 2012; Villadangos et al., 2010). Likewise, some mutant strains of C. glutamicum to obtain specific bioaccumulators of any of the inorganic forms of arsenic, either arsenite or arsenate (Villadangos et al., 2014) .In the absence of arsenite (the reduced form of inorganic arsenic), the regulator ArsR is repressing the expression of the rest of the genes; in its presence, ArsR has a high affinity for arsenite and is released from the operator, thus allowing the transcription of the two remaining genes (Villadangos et al., 2011). Intracellular arsenate reductase enzymes reduce arsenate to arsenite (the latter is very toxic), which is rapidly released to the outside of the cell by transmembrane arsenite permeases (Fu et al., 2009; Villadangos et al., 2012) (Fig. 2).
Currently, the main objective of the research is focused on the study of the differential antioxidant defense mechanisms that actinobacteria present, using as models C. glutamicum and Rhodococcus equi (intermediate model between C. glutamicum and M. tuberculosis). The most ubiquitous Redox system in cells is the thioredoxin (Trx)/thioredoxin reductase (TrxR) system, present in prokaryotes and eukaryotes and constituting a first line of defense against oxidation. However, evolutionary processes have allowed the appearance of other alternative Redox systems that are of a different nature depending on the organism in question. These alternative systems depend on specific redoxin and low molecular weight compounds. Our research group has pioneered the description of a new cellular Redox system exclusive to actinobacteria that includes the low molecular weight pseudosugar mycothiol (MSH) and the corresponding redoxin capable of specifically recognizing MSH; this redoxin was named mycorredoxin (Mrx) (Ordóñez et al., 2009). The “appearance” of this differential Redox buffer means that the MSH / Mrx pair and / or any of the proteins that interact with MSH / Mrx (mycotiolation) can constitute new antimicrobial targets that can be used specifically for this group of microorganisms ( Van laer et al., 2012; Van laer et al., 2013). Studies based on the degree of importance and / or essentiality of each of these targets currently constitute the bulk of our line of research.
During the last years our group is collaborating with other research groups that have common scientific interests, and led by researchers: William Margolin (USA), Klas Flardh (Sweden), Virginie Molle (France), Richard Daniel (UK), Joris Messens (Belgium), Joern Kalinowsky (Germany), Barry P. Rosen (USA), Juan Ayala (Madrid) and Jose A. Ainsa (Spain).
Fiuza M, Canova MJ, Patin D, Letek M, Zanella-Cleon I, Becchi M, Mateos LM, Mengin-Lecreulx D, Molle V, Gil JA. (2008a). The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum. J Biol Chem. 283:36553-36563.
Fiuza M, Canova MJ, Zanella-Cleon I, Becchi M, Cozzone AJ, Mateos LM, Kremer L, Gil JA, Molle V. (2008b). From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J Biol Chem. 283:18099-18112.
Fiuza M, Letek M, Leiba J, Villadangos AF, Vaquera J, Zanella-Cleón I, Mateos LM, Molle V, Gil JA. (2010). Phosphorylation of a novel cytoskeletal protein (RsmP) regulates rod-shape morphology in Corynebacterium glutamicum. J Biol Chem. 285:24387-24397.
Fu HL, Meng Y, Ordóñez E, Villadangos AF, Bhattacharjee H, Gil JA, Mateos LM, Rosen BP. (2009). Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. J Biol Chem. 284:19887-19895.
Letek M, Ordóñez E, Vaquera J, Margolin W, Flärdh K, Mateos LM, Gil JA. (2008). DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol. 190:3283-3292.
Letek M, Fiuza M, Villadangos AF, Mateos LM, Gil JA. (2012). Cytoskeletal proteins of actinobacteria. International. J Cell Biol. doi:10.1155/2012/905832.
Mateos LM, Ordóñez E, Letek M, Gil JA. (2006). Corynebacterium glutamicum as a model bacterium for the bioremediation of arsenic. Int Microbiol. 9:207-215.
Mateos LM, Letek M, Villadangos AF, Fiuza M, Ordóñez E, Gil JA. (2011). Chapter 6: Corynebacterium. In: Molecular detection of human bacterial pathogens. Edited by: Dongyou Liu. Taylor y Francis CRC Press. USA. ISBN: 978-1439812389.
Ordóñez E, Van Belle K, Roos G, De Galan S, Letek M, Gil JA, Wyns L, Mateos LM, Messens J. (2009). Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J Biol Chem. 284:15107-15116.
Ordóñez E, Villadangos AF, Fiuza M, Pereira FJ, Gil JA, Mateos LM, Aller AJ. (2012). Modelling of arsenate retention from aqueous solutions by living coryneform double-mutant bacteria. Environm Chem. 9:121-129.
Valbuena N, Letek M, Ordóñez E, Ayala J, Daniel RA, Gil JA, Mateos LM. (2007). Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol Microbiol. 66:643-657
Van Laer K, Buts L, Foloppe N, Vertommen D, Van Belle K, Wahni K, Roos G, Nilsson L, Mateos LM, Rawat M, van Nuland NA, Messens J. (2012). Mycoredoxin-1 is one of the missing links in the oxidative stress defence mechanism of Mycobacteria. Mol Microbiol. 86:787-804.
Van Laer K, Dziewulska AM, Fislage M, Wahni K, Hbeddou A, Collet JF, Versées W, Mateos LM, Tamu Dufe V, Messens J. (2013). NrdH-redoxin of Mycobacterium tuberculosis and Corynebacterium glutamicum dimerizes at high protein concentration and exclusively receives electrons from thioredoxin reductase. J Biol Chem. 15:7942-7955.
Villadangos AF, Ordóñez E, Muñoz MI, Pastrana IM, Fiuza M, Gil JA, Mateos LM, Aller AJ. (2010). Retention of arsenate using genetically modified coryneform bacteria and determination of arsenic in solid samples by ICP-MS. Talanta. 80:1421-1427.
Villadangos AF, Van Belle K, Wahni K, Dufe VT, Freitas S, Nur H, De Galan S, Gil JA, Collet JF, Mateos LM, Messens J. (2011). Corynebacterium glutamicum survives arsenic stress with arsenate reductases coupled to two distinct redox mechanisms. Mol Microbiol. 82:998-1014.
Villadangos AF, Fu HL, Gil JA, Messens J, Rosen BP, Mateos LM. (2012). Efflux permease CgAcr3-1 of Corynebacterium glutamicum is an arsenite-specific antiporter. J Biol Chem. 287:723-735.
Villadangos AF, Ordonez E, Pedre B, Messens J, Gil JA, Mateos LM. (2014). Engineered coryneform bacteria as a bio-tool for arsenic remediation. Appl Microbiol Biotechnol. DOI 10.1007/s00253-014-6055-2.
