
Group photo.Above: Carlos Serna, Emilia Wedel, Jose F. Delgado, Bruno González-Zorn, Bosco Rodríguez, Cristina Bernabé. Below: María Eugenia Revilla, Niloofar Razmgah, Mónica Suárez, Natalia Montero, Irene Sánchez. Quadrants: Gabriel Moyano, Manuel Ares, Ana Sempere, Cristina Calvo.
Antibiotic resistance (AMR) represents one of the greatest threats of the 21st century. Its progress entails a high cost, claiming more than 33,000 human lives per year in the European Union, a figure that is estimated at 10 M by the year 2050. Our group considers AMR as an ecological phenomenon whose foundation we approach from a One Health or One Health (Gonzalez-Zorn et al., 2013). Our objective is to try to understand, at a fundamental level, what is the flow of antibiotic resistance mechanisms in our Earth ecosystem, using different reductionist approaches. ARU was born in 2004 and during this time we have defended twelve doctoral theses. Our first doctorates, Álvaro San Millán and José Antonio Escudero, now have their own research group with two ERC Starting Grants, while others have joined the ECDC's training program of excellence or are doing their postdoc in the US or the UK . We have participated and continue to participate in relevant EU projects on antibiotic resistance, such as EvoTAR (Evolution and Transfer of Antimicrobial Resistance), EFFORT (Ecology from Farm to Fork of Resistance Transmission), EJP-OH (European Joint Program on One Health) or AVANT (Alternatives to Antimicrobials), in addition to two ITN Marie Skłodowska-Curie TRAIN-ASAP and CARTNET. Since our inception we have collaborated with the University for Development Studies in Ghana. We built a Food Safety laboratory there and trained Courage K.S. for four years. Saba, now in charge of it (Saba et al., 2012). With the help of WHO, the International Foundation for Science and UCM, we have established a blood culture system and conducted microbiology and biotechnology workshops at the Tamale Teaching Hospital. Every year we send students with UCM and CAM programs to investigate, train and volunteer. We are part of the NEAR-JPIAMR Network with other EU and African countries to assist in their AMR research. In our early days, we identified a hitherto unknown resistance mechanism in Escherichia coli called armA (aminoglycoside resistance methyltransferase), a 16S RNA methyltransferase that nullifies the effect of aminoglycosides (González-Zorn et al., 2005). We characterized how this resistance mechanism acts at the ribosome level exclusively on the m7 G1405 residue, interfering with the ribosome's own methylome and with global translation in the bacterium itself (Gutiérrez et al., 2013). These genes confer high levels of resistance to temamycin, a last-resort antibiotic approved in the US in 2018, in the approval phase for clinical use in Europe. Therefore, it is essential that we continue to work on 16S RNA methyltransferases if we want a new antibiotic to be effective for as long as possible. During these years, we have also identified that small multicopy plasmids (MCPs) have much more relevance than they have been given in recent decades. In the group we have discovered that MCPs are present in a large number of pathogenic and non-pathogenic bacteria (Ares-Arroyo 2018). These MCPs capture resistance mechanisms and spread them with a low fitness cost, which is fully compensated through chromosomal mutations (San MiIllán 2015). We are fascinated by how these MCPs enable genetic innovation giving rise to a new family plasmid with its own copy number. The discovery of the relevance of MCPs has created multiple lines of research that open new doors in the study of plasmids and their relevance in AMR. These previous studies led us to be interested in genetic uptake and transmission. In collaboration with other groups, we discovered the implication of the SOS mechanism in the uptake of resistance cassettes in bacterial integrons (Guerin 2009) and we continue to investigate this mechanism as an element of regulation of AMR and the plasmid mechanisms involved in its dissemination (Hadziabdic S 2018). Within this line of research, we discovered that the healthy population of large cities represents an important reservoir of emerging resistance mechanisms, such as mcr-1-mediated resistance to colistin (Martínez-Ovejero C 2017). This discovery earned us the PRAN National Research Award in 2018, merit of all ARU members and especially Jose F. Delgado-Blas. In addition, we are currently using mouse models to understand plasmid transmission and antibiotic resistance mechanisms between individuals using video-tracking systems. If we have to highlight something during the existence of ARU, it is undoubtedly the human level of all the members of the group. The support between them gives rise to a cohesion and camaraderie that we consider unique and essential. We have also felt the support of all our colleagues from the Molecular Microbiology Group, from the entire SEM and from the Spanish microbiological community, thanks to which we settled as a group in Spain. To all, thank you very much.
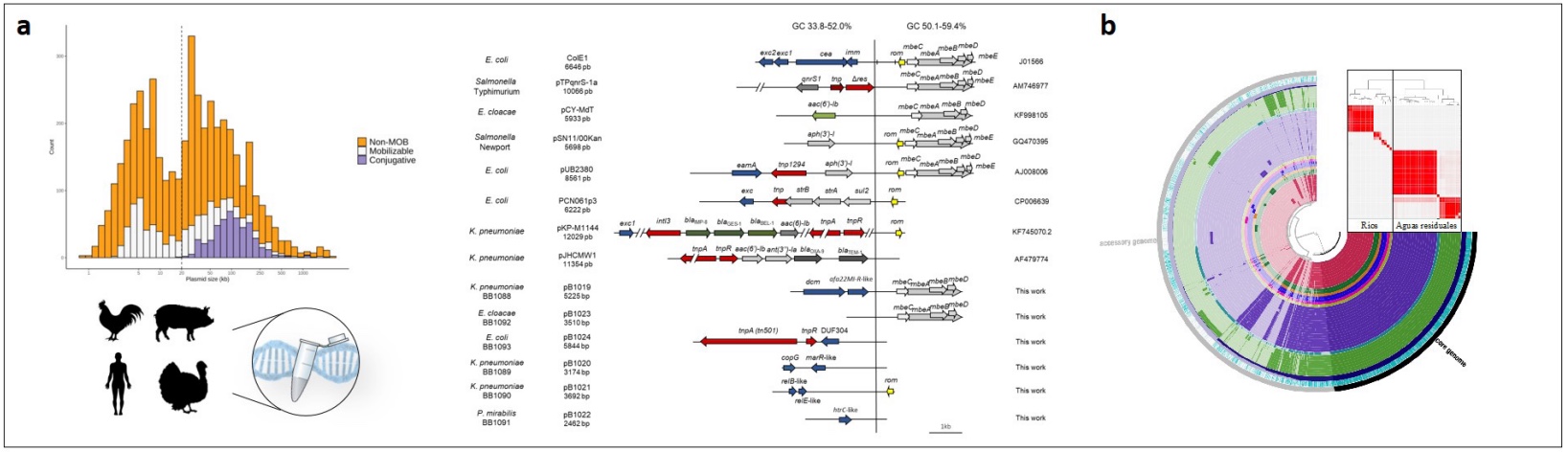
Figure 1. to. Discovery of the relevance of multicopy plasmids (MCPs). Using classical techniques, together with genomics and bioinformatics in humans, animals and the environment, we have revealed the enormous relevance of small plasmids in microbiology. Almost half of the plasmids in the databases are small in size. (Ares–Arroyo, 2018). b. The comparative analysis of the pangenome of E. coli isolates carrying 16S rRNA methyltransferase genes from natural effluents and wastewater allows us to establish links between bacteria, environmental mobilomas, the anthropogenic effect and its evolution over time.
Representative bibliography
Gonzalez-Zorn B, Catalan A, Escudero JA, Domínguez L, Teshager T, Porrero C, Moreno MA. Genetic basis for dissemination of armA. J Antimicrob Chemother. 2005 Sep;56(3):583-5. Epub 2005 Jul 18.
Guerin, G. Cambray, N. Sanchez-Alberola, S. Campoy, I. Erill, S. Da Re, B. Gonzalez-Zorn, J. Barbe, M. C. Ploy, D. Mazel. Recombination of integron cassettes is under control of the SOS response. Science 324(5930):1034. 2009.
Saba C.K.S. and B. Gonzalez-Zorn. Food Safety in Ghana: A Metha-Analysis. J Infect Dev Ctries 6(12):828-35. 2012.
González-Zorn B, Escudero JA. Ecology of antimicrobial resistance: humans, animals, food and environment. Int Microbiol. 15(3):101-9. Review. 2013.
Gutiérrez B, Douthwaite S, Gonzalez-Zorn B. Indigenous and acquired modifications in the aminoglycoside binding sites of Pseudomonas aeruginosa rRNAs. RNA Biol. 10(8):1324-32. 2013.
San Millan A, Santos-Lopez A, Ortega-Huedo R, Bernabe-Balas C, Kennedy SP, Gonzalez-Zorn B. Small plasmid-mediated antibiotic resistance in Haemophilus influenza is enhanced by increases in plasmid copy number and bacterial fitness. Antimicrobial Agents Chemotherapy. 59(6):3335-41. 2015.
Ovejero CM, Delgado-Blas JF, Calero-Caceres W, Muniesa M, Gonzalez-Zorn B. Spread of mcr-1 –carrying Enterobacteriaceae in sewage water from Spain. J Antimicrob Chemother. 2017 Jan 10. pii: dkw533. doi: 10.1093/jac/dkw533.
Hadziabdic S, Fischer J, Malorny B, Borowiak M, Guerra B, Kaesbohrer A, Gonzalez-Zorn B, Szabo I. In vivo Transfer and Microevolution of Avian Native IncA/C2 blaNDM-1-Carrying Plasmid pRH-1238 during a Broiler Chicken Infection Study.Antimicrob Agents Chemother. 2018 Mar 27;62(4). Pii: e02128-17. Doi: 10.1128/AAC.02128-17.
Ares-Arroyo M, Bernabe-Balas C, Santos-Lopez A, Baquero MR, Prasad KN, Cid D, Martin-Espada C, San Millan A, Gonzalez-Zorn B. PCR-Based Analysis of ColE1 Plasmids in Clinical Isolates and Metagenomic Samples Reveals Their Importance as Gene Capture Platforms. Front Microbiol. 2018 9:469.
Munk P et al. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat Microbiol. 2018 Oct. 3 (10):1186. Doi: 10.1038/s41564-018-0241-4.
