
Group photo. From left to right: Jon Sicilia, Jerónimo Rodríguez, Carmen de la Vega, Cristina Herencias, Álvaro San Millán, Aida Alonso, Ricardo León, Javier de la Fuente and Laura Toribio.
The group Plasmid Biology and Evolution (PBE lab, www.pbelab.es) was born in January 2016 and since then it has grown steadily until reaching its current composition (Figure 1). The PBE lab belongs to the Microbiology, Immunology and Infection Area of the Ramón y Cajal Institute for Health Research, at the Ramón y Cajal University Hospital (Madrid). At the PBE lab, our main interest is the study of the evolutionary dynamics of plasmids in bacterial populations. Plasmids are genetic elements that replicate independently of the chromosome and are capable of transferring genetic information horizontally between bacteria in a process known as conjugation. In this way, plasmids play a key role in bacterial evolution and ecology because they allow bacteria to access new adaptive traits. The most illustrative example of the ability of plasmids to enhance bacterial evolution is the key role they have played in the dissemination of antibiotic resistance mechanisms. This phenomenon has strategically contributed to the development of the global crisis of antibiotic resistance that we currently face (MacLean and San Millan, 2019).
In our group we mainly develop two main lines of research:
Real-time evolution of antibiotic resistance in the hospital
Conjugative plasmids play an essential role in the spread of antibiotic resistance in scenarios of clinical relevance (San Millan, 2018). One of the main factors that compromises the success of the dissemination of conjugative plasmids is the biological cost they impose on their bacterial hosts, which translates into a decrease in the reproductive capacity of the bacteria carrying the plasmid (San Millan and MacLean, 2017). The origin of these costs, and the compensatory evolution responsible for mitigating them, remain largely unknown. These evolutionary dynamics are extremely relevant, because they will be at least partially responsible for the appearance of successful bacteria-plasmid associations that spread uncontrollably in hospitals (San Millan, 2018).
In our group we try to understand the genetic and evolutionary bases that determine the establishment of these successful associations in the hospital. To do this, and thanks to the collaboration with the hospital's excellent clinical microbiologists, we studied the evolution of Enterobacteriaceae strains carrying antibiotic resistance plasmids isolated from the intestinal microbiota of hospitalized patients (Figure 1). We combine genomics, transcriptomics, gene editing with CRISPR technology, CRISPRi screening and flow cytometry techniques with mathematical modeling to characterize the epidemiology and evolution of bacteria carrying antibiotic resistance plasmids (DelaFuente et al., 2020; San Millan et al. , 2014b; San Millan et al., 2018; San Millan et al., 2015). Our results help predict, and potentially contain, the dissemination of bacteria-plasmid associations of clinical interest in our hospital.
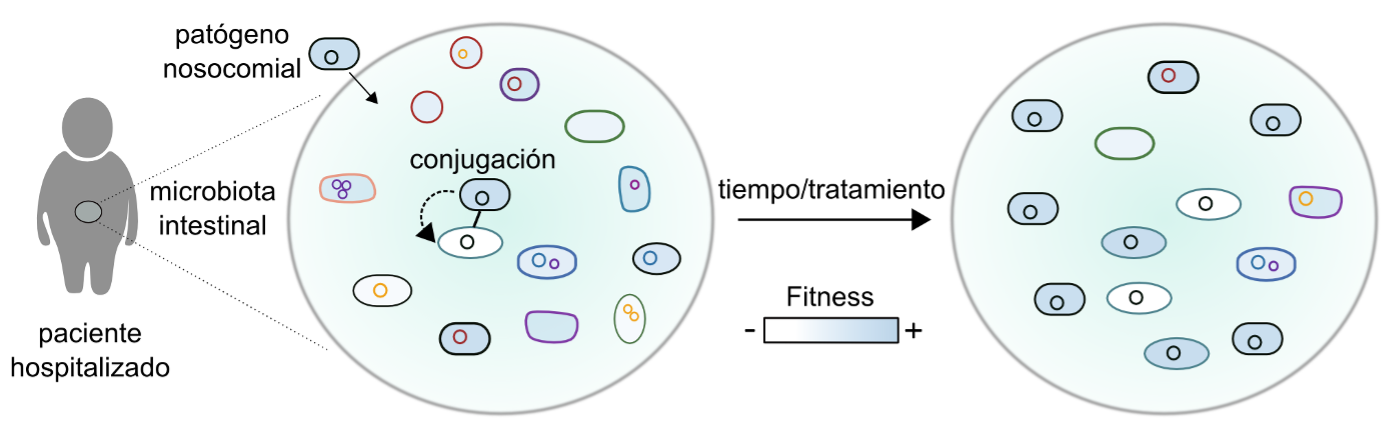
Figure 1. Evolution of plasmid-mediated antibiotic resistance. Modified from (San Millan, 2018). Evolutionary dynamics of bacteria-plasmid associations in the intestinal microbiota of a hospitalized patient. A nosocomial pathogen carrying an antibiotic resistance plasmid colonizes the intestinal microbiota and transfers the plasmid to resident bacteria in this microbiota. The plasmid can produce a biological cost (i.e. reduced fitness. Legend: white, low fitness; blue, high fitness), but is selected for by antibiotic treatment. Over time, this cost disappears thanks to compensatory mutations.
Role of multicopy plasmids in bacterial evolution
Plasmids can be broadly divided into those of large size and low copy number and those of small size and high copy number. Although the size limit varies between bacterial families, this bimodal distribution of sizes and copies remains constant throughout the phylogeny (San Millan et al., 2014a; Smillie et al., 2010). Large conjugative plasmids play a decisive role in the spread of antibiotic resistance and have therefore been extensively studied in recent decades. Small multicopy plasmids are extremely common in prokaryotes, but their role in bacterial biology and evolution has been less well studied.
In our laboratory we investigate the possible evolutionary advantages associated with multicopy plasmids. One of the most outstanding features of these plasmids is the fact that they represent an island of polyploidy in the haploid genome of most bacteria. In recent studies, we have shown that the polyploidy provided by these plasmids has at least two important consequences: (i) accelerate the evolution of the genes they encode (San Millan et al., 2016) and (ii) maintain the allelic diversity of the plasmids. same in the host bacteria (Rodriguez-Beltran et al., 2018) (Figure 2).
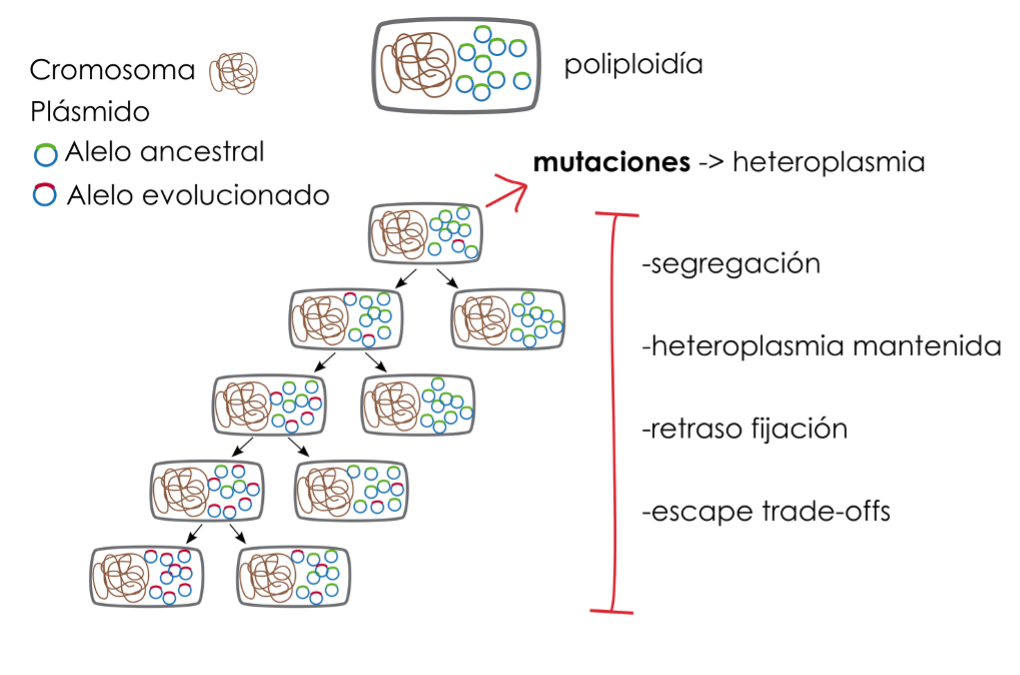
Figure 2. Evolutionary dynamics of multicopy plasmids. The polyploidy associated with multicopy plasmids increases the chances of acquiring mutations in their genes. The coexistence of versions of the same plasmid with and without the mutation is known as heteroplasmy. The replication and segregation dynamics of these plasmids delay the fixation of mutations. Maintained heteroplasmy provides bacteria with allelic diversity, which may have adaptive value by allowing bacteria to escape trade-offs between the different activities of gene alleles.
In summary, in our group we try to understand the population biology of antibiotic resistance plasmids by combining advanced techniques of molecular biology and evolutionary biology. Our ultimate goal is to apply the concepts we learn from studying the evolution of plasmid-mediated antibiotic resistance to the development of more rational strategies for the control of infectious diseases.
Representative bibliography
DelaFuente, J., Rodriguez-Beltran, J., San Millan, A., 2020. Methods to Study Fitness and Compensatory Adaptation in Plasmid-Carrying Bacteria. Methods Mol Biol 2075, 371-382.
MacLean, R.C., San Millan, A., 2019. The evolution of antibiotic resistance. Science 365, 1082-1083.
Rodriguez-Beltran, J., Hernandez-Beltran, J.C.R., DelaFuente, J., Escudero, J.A., Fuentes-Hernandez, A., MacLean, R.C., Pena-Miller, R., San Millan, A., 2018. Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nat Ecol Evol 2, 873-881.
San Millan, A., 2018. Evolution of Plasmid-Mediated Antibiotic Resistance in the Clinical Context. Trends Microbiol 26, 978-985.
San Millan, A., Escudero, J.A., Gifford, D.R., Mazel, D., MacLean, R.C., 2016. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nature Ecology & Evolution 1, 0010.
San Millan, A., Heilbron, K., MacLean, R.C., 2014a. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J 8, 601-612.
San Millan, A., MacLean, R.C., 2017. Fitness Costs of Plasmids: a Limit to Plasmid Transmission. Microbiology Spectrum 5.
San Millan, A., Peña-Miller, R., Toll-Riera, M., Halbert, Z.V., McLean, A.R., Cooper, B.S., MacLean, R.C., 2014b. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat Commun 5, 5208.
San Millan, A., Toll-Riera, M., Qi, Q., Betts, A., Hopkinson, R.J., McCullagh, J., MacLean, R.C., 2018. Integrative analysis of fitness and metabolic effects of plasmids in Pseudomonas aeruginosa PAO1. ISME J 12, 3014-3024.
San Millan, A., Toll-Riera, M., Qi, Q., MacLean, R.C., 2015. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat Commun 6, 6845.
Smillie, C., Garcillán-Barcia, M.P., Francia, M.V., Rocha, E.P., de la Cruz, F., 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74, 434-452.
