
Group photo. Jose Ramos Alive
The Cellular Microbiology laboratory of the IDIVAL Institute has a recent history. It really started in 2009, with the arrival in Santander of the Principal Investigator, but it was at the end of 2012 when the group completed the equipping of its 3 small laboratories, one for Molecular Biology, another for Cell Biology and one for Microbiology, with funds from of national and international projects. The main line of research of our group focuses on the study of pathogen-host interactions, using models involving pathogenic bacteria and in vitro cell and tissue cultures (mouse-rat-human). Among the pathogenic bacteria we studied are Listeria monocytogenes, Staphylococcus aureus Listeria monocytogenes, Staphylococcus aureus Serratia Listeria monocytogenes, Staphylococcus aureus AcinetobacterListeria monocytogenes, Staphylococcus aureus Listeria monocytogenes, Staphylococcus aureusListeria monocytogenes, Staphylococcus aureus Acinetobacter They are very important in hospital infections, especially in intensive care units (ICUs) since they have a propensity to develop resistance to multiple classes of antibiotics, which reduces therapeutic options to combat them. While the epidemiology and antibiotic resistance of most of these species are being extensively studied, the molecular and genetic basis of their virulence remain largely unexplored. In addition, there is a great lack of knowledge of host responses to these pathogens. Acinetobacter It is currently a serious health problem in many Spanish hospitals and is therefore one of the pathogens of interest for the Spanish Network for Infectious Pathology (REIPI) to which our group belongs. Responding to the need for studies on the mechanisms involved in the host-pathogen (HP) relationship, our team develops multidisciplinary research on HP interactions mainly in these species, although we have also started work in the field of HP interactions with probiotic strains, thus initiating different collaborations. Some examples of these interactions can be seen in Figure 1.
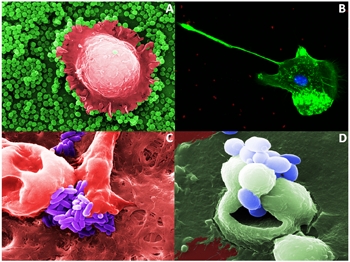
Figure 1. A. Scanning microscopy (SEM). Macrophage phagocytizing a Staphylococcus aureus biofilm. Magnification ×10,000. b. Fluorescence microscopy. Human macrophage capturing Acinetobacter baumannii (red). Magnification ×400. c. Scanning microscopy (SEM). Brain macrophages phagocytizing Listeria monocytogenes. Reproduced with permission from John Wiley and Sons Publishers: GLIA, 2013; Vol 61:611-22. Magnification ×20,000. d. Scanning microscopy (SEM). Yeasts (blue) invading a human epithelial cell. Magnification ×10,000
The objectives of the team are: A) To develop new tools for the in-depth study of bacteria-cell interactions; B) Study the impact of different antibiotics during H-P interactions in vitro; C) Carry out a detailed analysis of the expression of genes of immune cells in response to different phenotypes of these multiresistant species or their bacterial products, as well as the expression of virulence genes in pathogenic bacteria.
By using Microbiology, Cell Biology and Advanced Microscopy techniques, the models we use are very flexible and can be adapted to the study of any human pathogen. Specifically, with our national and international collaborations, we have already studied 5 of the 6 multi-resistant ESKAPE pathogens (By using Microbiology, Cell Biology and Advanced Microscopy techniques, the models we use are very flexible and can be adapted to the study of any human pathogen. Specifically, with our national and international collaborations, we have already studied 5 of the 6 multi-resistant ESKAPE pathogens ( and By using Microbiology, Cell Biology and Advanced Microscopy techniques, the models we use are very flexible and can be adapted to the study of any human pathogen. Specifically, with our national and international collaborations, we have already studied 5 of the 6 multi-resistant ESKAPE pathogens (By using Microbiology, Cell Biology and Advanced Microscopy techniques, the models we use are very flexible and can be adapted to the study of any human pathogen. Specifically, with our national and international collaborations, we have already studied 5 of the 6 multi-resistant ESKAPE pathogens (
By using Microbiology, Cell Biology and Advanced Microscopy techniques, the models we use are very flexible and can be adapted to the study of any human pathogen. Specifically, with our national and international collaborations, we have already studied 5 of the 6 multi-resistant ESKAPE pathogens (
Representative bibliography
By using Microbiology, Cell Biology and Advanced Microscopy techniques, the models we use are very flexible and can be adapted to the study of any human pathogen. Specifically, with our national and international collaborations, we have already studied 5 of the 6 multi-resistant ESKAPE pathogens ( New aspects in the biology of Photobacterium damselae subsp. PiscicidesNew aspects in the biology of
New aspects in the biology of New aspects in the biology of Streptococcus iniae New aspects in the biology of
New aspects in the biology of New aspects in the biology of Rhodococcus equi New aspects in the biology of
New aspects in the biology of New aspects in the biology of Serratia dissolving New aspects in the biology of
Remuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J. Remuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J. Listeria monocytogenesRemuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J.
Remuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J. Remuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J. Listeria monocytogenesRemuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J.
Remuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J. Remuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J. Hafnia alvei and Hafnia paralveiRemuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J.
