
Group photo. EXOPOL team
EXOPOL S.L., is a spinoff of the University of Zaragoza with more than 20 years of experience in diagnosis and production of autovaccines in animal health; areas to which it dedicates more than 50% of its R+D+i effort and in which it maintains collaborations with more than 25 research groups at a national and international level. Throughout its history, the company has obtained financing to carry carried out various research projects (CDTI) aimed mainly at improving the current diagnosis and the production of vaccines in the veterinary field.
EXOPOL's extensive experience in these areas, including real-time PCR (qPCR) molecular diagnostics, has enabled the company to design and validate its own assays, which it has designated as “EXOone qPCREXOPOL's extensive experience in these areas, including real-time PCR (qPCR) molecular diagnostics, has enabled the company to design and validate its own assays, which it has designated as “ EXOone qPCR, makes use of a new technology to stabilize all the reagents necessary for the reaction in the wells of a qPCR plate or strips, which has made it possible to obtain a "ready-to-use" product, with storage at 4º C, in addition to other advantages .The stabilization also allows these qPCR kits to be stored at refrigeration temperatures (+4°C to +8°C) without the need to freeze them; which is a great practical advantage since it facilitates transport and avoids the tedious thawing process of the different reagents necessary to prepare a mixture in a common qPCR assay.
Another important feature of the essays EXOone qPCR is that they have all been designed to use the same thermal protocol; which allows the possibility of evaluating several pathogens in the same amplification run, reducing costs and resources. This advantage of our tests allows us to design specific diagnostic panels to suit the client, such as the one shown below.
Currently, the company continues to design new products and seek collaborations with research groups interested in applying this technology in other areas of molecular diagnosis such as human health, agri-food or the environment, among others.
recently described Burd et al 2010 (1) y Saunders et al 2013 (2). At EXOPOL we have followed these criteria in order to provide a quality product that is both quick and easy to use.
Assay validation results EXOone qPCR for the identification of relevant pathogens in our sector such as: Coxiella burnetii (3), Pathogenic Leptospira (4), Toxoplasma gondii (5), Mycoplasma agalactiae (6), Mycoplasma bovis (7), etc; They have been presented at several highly relevant national and international conferences such as WAVLD, EAVLD, AVEDILA, SEM, among others. Additionally, other qPCR assays have been developed for the identification of bacterial, parasitic or viral pathogens of veterinary interest, the results of which are currently in the process of being published (for a detailed list visit: www.exopol.com).
Assay validation EXOone qPCR Assay validation
– Assay validation Assay validation Assay validation Assay validation1 Assay validationAssay validation Assay validation
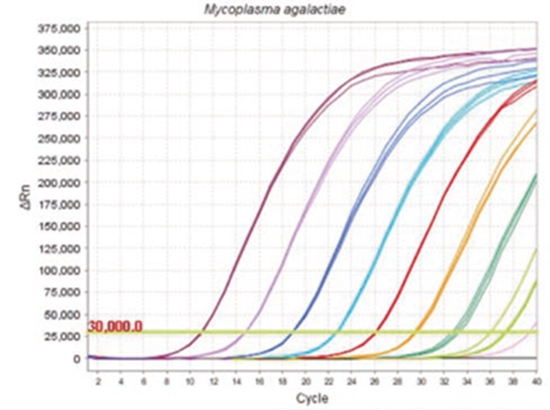
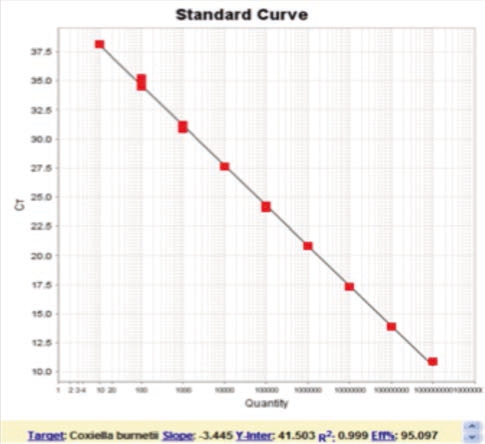
Fig.2 Assay validation
– Concordance studies: By evaluating a panel of reference strains, vaccine strains, field strains, and positive and negative clinical cases for each pathogen; which are evaluated in parallel with the most important commercial qPCR assays on the market (LSI, Adiagen, Ingenetix, etc.) to determine the level of agreement. The essays EXOone qPCR have been shown to have an excellent concordance (Kappa 0.87 to 1.0) with the qPCR kits offered by the relevant companies in our sector.
- Studies specificity: In a study that includes the evaluation of a panel of between 25 to 45 microorganisms (bacteria, viruses and parasites) that are genetically related, that cause related diseases or co-infections, or that can be found as natural microbiota in the selected samples.
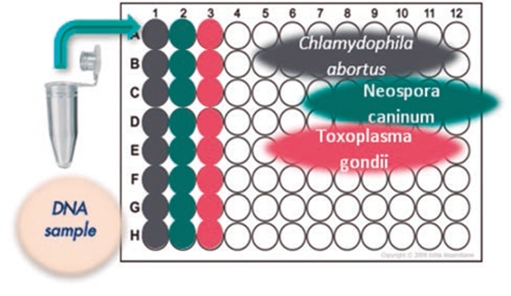
Fig.3 Reproductive panel for the simultaneous evaluation (multiparametric) of Chlamydophila abortus, Neospora caninum and Toxoplasma gondii.
Key Features of the EXOone qPCR Assays
The new technology used in the trials EXOone qPCR, by stabilizing all the necessary reagents in wells, allows faster qPCR setup; since it only needs the addition of DEPC water (supplied with the kit) and the nucleic acid sample, before taking the reaction to the thermocycler. This makes the work of the technician much easier and also avoids possible pipetting errors during the preparation of the assay, with the consequent improvement in its repeatability and reproducibility.
In summary, the stabilized assay EXOone qPCR, due to their “ready to use” format and their careful validation, they are an excellent diagnostic tool for the identification or quantification of pathogens of veterinary interest, standing out not only for their sensitivity and specificity but also for their ease of use.
Representative bibliography
Benito AA, Arnal JL, de Tomas E, et al. Internal controls for reliable results in real-time pcr diagnostic assays. WAVLD, Berlin 2013.
Benito AA, Villa A, Serrano JD, et al. Development of a Real Time PCR for the diagnosis of Toxoplasma gondii. SEM IX Molecular Microbiology group meeting. Majorca, 2012.
Burd Eileen M. Validation of Laboratory-Developed Molecular Assays for Infectious Diseases. Clinical Microbiology Reviews 2010. Vol.23 (3):
Saunders N, Zambon M, Sharop I, et al. (2013). Guidance on the development and validation of diagnostic tests that depend on nucleic acid amplification and detection. Journal of Clinical Virology 56
Villa A, Arnal JL, Serrano DJ, et al. Development and validation of a Real-Time PCR Zen Gel Mix for the diagnosis and quantification of Coxiella burnetii. EAVLD, Poland 2012a.
Villa A, Benito AA, Bernal JL et al. Leptospirosis: Diagnosis by real-time PCR and microbiological isolation. AVEDILA, Badajoz.2012b.
Villa A, De Tomás E, Benito AA, Arnal JL, Serrano JD. Evaluation of cloned genes as quantitative controls in Real-Time assays for infectious diseases with veterinary importance. SEBBM 2012c, Sevilla.
Villa A, Fernandez A, JL Arnal, et al. Development and validation of a ZEN Real Time PCR Gel Mix for the detection and quantification of the gene (mp81) of Mycoplasma agalactiae. AVEDILA, Tenerife 2011.
Villa A, Serrano JD, Benito AA, et al. Targeting uvrC gene for Mycoplasma bovis Gel Mix Real Time PCR. WAVLD, Berlin 2013.
