
Group photo. Members of the group "Structure, Dynamics and Function of Rhizobacteria Genomes" today.
Our group "Structure, Dynamics and Function of Rhizobacteria Genomes" aims to obtain basic and applied knowledge about the beneficial interactions between plants and microorganisms of interest in agricultural and forestry systems (https://www.eez.csic.es/ dynamic-structure-and-function-of-rhizobacteria-genomes). The group was constituted at the end of the 90's as a drift of the lines within the Department of Soil Microbiology and Symbiotic Systems in our center - La Estación Experimental del Zaidín – based on the study of symbiosis Rhizobium-legume. The group is made up of four researchers on staff: Nicolás Toro, Francisco Martínez-Abarca, Manuel Fernández López and Jose I. Jiménez Zurdo. It currently has a total of 14 members including technical staff, Master's and Predoctoral students as well as Postdoctoral staff. He focuses his lines of research on the ecology of rhizospheric microorganisms and their use in the recovery of degraded soils, in the genomic analysis of soil microbial communities, the characterization of new regulatory RNAs in symbiotic microorganisms and finally in the contribution to development of functional genomics of microorganisms and plants through the use of high-throughput mutagenesis tools. This latest line of research (@ToroLaboratory) was developed based on discoveries made at the beginning on a particular isolate, GR4, of the bacterium Sinorhizobium meliloti.
During the 1980s, the study of the genetic determinants causing greater efficiency in plant nodulation led us to a specific 40 kb zone contained in one of the two cryptic plasmids present in this isolate (pRmeGR4b). This point is the beginning of the first key shift in research within the group that led us to become interested in a mobile element present in this region that has given rise to the development of an entire on-demand mutagenesis technology derived from what is barely known. known at the time (late 1990s) as group II introns. In this region, a locus of 1894 nt was found containing the first active retroelement in rhizobacteria: RmInt1 (Rm of Rhizobium meliloti, Int1, the first Intron). Group II introns are large ribozymes that behave like mobile elements that undergo "splicing" through a mechanism reminiscent of eukaryotic introns. Its mobility is mediated by the formation of a ribonucleoprotein complex made up of the protein encoded by the intron itself and the excised RNA. The target DNA it is directed to is recognized primarily by RNA that can be efficiently programmed to any target DNA on demand.
Successive years of work with this element laid the foundations for new varieties of introns with which to develop targetron technology, giving rise to various research projects, as well as various international collaborations. A relevant activity present in this retroelement and necessary for its activity as a mutagenic tool is the Reverso Transcriptase (RT) activity. The presence and success of this retroelement in S. meliloti, has made us wonder if its presence at a certain time could have given evolutionary advantages to the microorganisms that contain it. Thus, new related elements, fragments of old elements inserted in critical areas conserved in the genomes of related rhizobia have appeared in new sequenced genomes. To this has also been added the effort and knowledge of the complete genome of GR4, the starting bacterium, which, thanks to our knowledge of this repeated element, allowed us to close it precisely, as well as tackle the sequencing of new variants that has allowed us to know the evolution times of this bacterium and its very likely association with the crop on which it depends (in this case, alfalfa).
The new Microbiology has changed the way we ask ourselves questions about the role of introns and other elements evolutionarily related to them in the hosts that contain them. Going one step further, we have been especially interested in understanding the origin of the RT present in this type of retroelements, for which, since 2014, we have been interested in knowing the phylogenetic relationships of the different types of reverse transcriptases present in the prokaryotic world. Since that year, we have observed a particular subgroup closely related to those present in group II introns that seemed to have completely changed their functional system. These reverse transcriptases were found to be part of the CRISPR locus, specifically associated mainly with genes of the adaptive complex, and in some cases forming a fusion protein (RT-Cas1). CRISPR-Cas systems constitute an "immune" defense system of bacteria against viruses and exogenous DNA. In the last phase of the process, a ribonucleoprotein (known as cas9) capable of recognizing “in vivo” a target sequence using a guide RNA and digesting it quickly, efficiently and precisely. This property has given rise to the so-called 'CRISPR Technology' for on-demand genome editing. A new question arises in our research group: What is the role of reverse transcriptases in CRISPR-Cas systems? What systems are they associated with? What type of co-evolution do they seem to have had with respect to the CRISPR-Cas system to which they are associated? We and other groups have found new varieties of RTs closely related to some CRISPR-Cas systems. In a recent work we have studied a new functional RT-CRISPR-Cas system. In our work we have demonstrated the essential role of reverse transcriptase activity in the spacer acquisition process. Process that in turn lacks sequence biases previously described in other systems. The study provides new tools with differential potential for the development of new biotechnological tools.
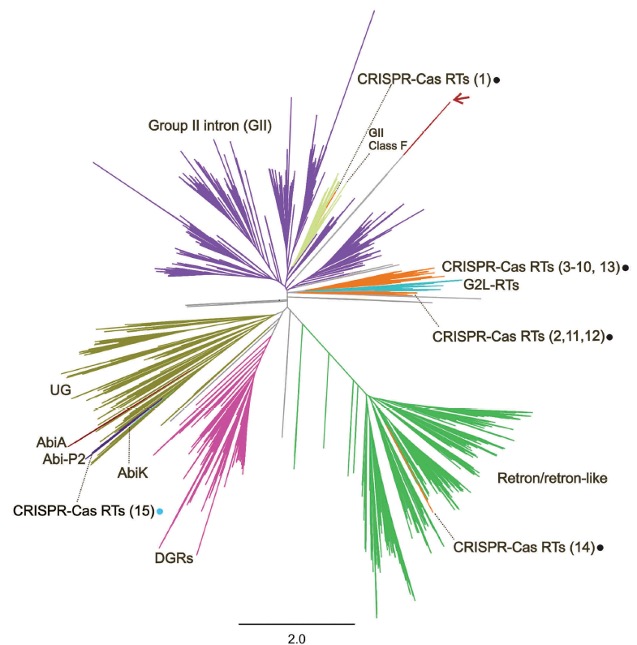
Figure 1. Phylogenetic tree that represents the prokaryotic diversity of Reverse Transcriptases (RTs) identified to date. Those that are closely related to CRISPR-Cas systems are highlighted with dots. (Modified from Toro et al. 2019, RNA Biol.
Most relevant recent publications
González-Delgado A, Mestre MR, Martínez-Abarca F, Toro N. (2019) Spacer acquisition from RNA mediated by a natural reverse transcriptase-Cas1 fusion protein associated with a type III-D CRISPR-Cas system in Vibrio vulnificus. Nucleic Acids Res. In press.
Toro N, Martínez-Abarca F, Mestre MR, González-Delgado A. (2019) Multiple origins of reverse transcriptases linked to CRISPR-Cas systems. RNA Biol. 16:1486-1493.
García-Rodríguez FM, Neira JL, Marcia M, Molina-Sánchez MD, Toro N. (2019) A group II intron-encoded protein interacts with the cellular replicative machinery through the β-sliding clamp. Nucleic Acids Res. 47:7605-7617.
Toro N, Martínez-Abarca F, Molina-Sánchez MD, García-Rodríguez FM, Nisa-Martínez R. (2018) Contribution of Mobile Group II Introns to Sinorhizobium meliloti Genome Evolution. Front Microbiol. 9:627.
Toro N, Martínez-Abarca F, González-Delgado A. (2017) The Reverse Transcriptases Associated with CRISPR-Cas Systems. Sci Rep. 7(1):7089.
Toro N, Martínez-Abarca F, Fernández-López M. (2016) The early events underlying genome evolution in a localized Sinorhizobium meliloti population. BMC Genomics.17:556.
Bull N, Nisa-Martinez R. (2014) Comprehensive phylogenetic analysis of bacterial reverse transcriptases. PLoS One. 9(11):e114083.
Toro N, Martínez-Rodríguez L, Martínez-Abarca F. (2014) Insights into the history of a bacterial group II intron remnant from the genomes of the nitrogen-fixing symbionts Sinorhizobium meliloti and Sinorhizobium medicae. Heredity (Edinb).113(4):306-15.
García-Rodríguez FM, Hernández-Gutiérrez T, Díaz-Prado V, Toro N. (2014) Use of the computer-retargeted group II intron RmInt1 of Sinorhizobium meliloti for gene targeting. RNA Biol. 11(4):391-401.
Martínez-Abarca F, Martínez-Rodríguez L, López-Contreras JA, Jiménez-Zurdo JI, Toro N. (2013) Complete Genome Sequence of the Alfalfa Symbiont Sinorhizobium/Ensifer meliloti Strain GR4. Genome Announc. 1(1). pii: e00174-12.
