
Group photo. From left to right: Ramón Santamaría, Margarita Díaz, Sergio Antoraz, Hector Rodríguez and David Sanz.
The research group led by doctors Ramón Santamaría and Margarita Díaz has focused its research on different aspects of the biology of bacteria of the genus Streptomyces. There are three main aspects that attract us to these microorganisms: 1) they produce a wide variety of antibiotics and antitumors; 2) they are normal inhabitants of the soil where they interact with other organisms and participate in the decomposition of organic matter; 3) they produce large amounts of hydrolytic enzymes, some of them with industrial potential.
To date, the most important interest of microorganisms of the genus Streptomyces, with more than 500 described species, is its ability to produce antibiotics and antitumor drugs (they produce more than 50% of all available natural antibiotics). This potential has been increased after sequencing more than 150 species and observing that all of them have multiple clusters biosynthetics that, inactive under laboratory conditions, are involved in the synthesis of unknown secondary metabolites. Achieving the production and identification of these cryptic metabolites and achieving adequate production levels, even of already known metabolites, is a task in which many groups and companies are involved worldwide using very varied approaches. Obtaining new bioactive compounds that deal with antibiotic resistance problems is one of the most important challenges of this century.
We are facing this challenge using two different strategies. On the one hand, we have focused on the study of regulation that different two-component systems exert on the production of antibiotics in order to be able to manipulate this regulation and enhance production routes of known antibiotics and / or activate silenced production routes. In this case, we use S. coelicolor as a model, and we have already studied the effect of six two-component systems on the production of antibiotics through the mutation of the genes that comprise them (Yepes et al. 2011). We are currently delving into the study of the three that have the most effect, and we have described that the systems we call AbrA1 / A2 and AbrB1 / B2 exert a negative effect on the production of antibiotics (Rico et al. 2014a, Sanz et al. Unpublished results) while the AbrC1 / C2 / C3 system (atypical system, composed of two kinases and a regulator) exerts a positive effect on the production of antibiotics. The use of the strains deleted in the negative systems as host strains for the heterologous expression of different antitumor pathways has allowed us to demonstrate their usefulness by doubling their production with respect to the parental strain (Rico et al. 2014a). Furthermore, the overexpression of the positive regulator AbrC3 in other species of Streptomyces induces the production of endogenous metabolites in them and can help us discover new bioactive molecules of interest for medicine produced by different species (Rico et al. 2014b and unpublished results).
On the other hand, we have focused on the effect exerted by the interaction of microorganisms of the genus Streptomyces with other soil microorganisms on the production of known antibiotics and on the activation of silenced pathways.
Our working hypothesis, together with that of other groups, is that many of the biosynthesis pathways present in the genomes of the Streptomyces that are silenced under laboratory conditions, could be activated in response to interaction with other organisms through molecular signaling between them in a way similar to that which should occur in the natural habitat.In fact, the study that we have initiated shows that the stimulation of the production of some antibiotics actually occurs in co-cultures of Streptomyces with other microorganisms such as Myxococcus O Bacillus (Pérez et al. 2011). Identifying the molecules that induce this effect is of great interest since it will allow their subsequent use in the means of production for the induction of these routes without the need to resort to co-cultures.
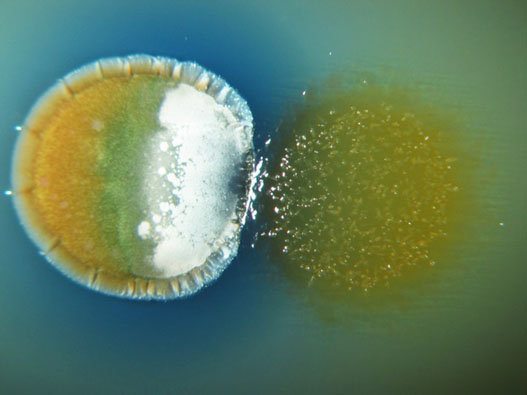
Figure 1. Interaction between Streptomyces coelicolor (left) and Myxococcus xanthus (right). Actinorhodin production is stimulated in the interaction zone (blue color).
The third line of work of the group focuses on the production capacity of secreted hydrolytic enzymes that these microorganisms possess and that allow them to degrade the organic matter of their main biological niche, the soil, and use the generated nutrients for their development. This secretory capacity, among other characteristics, makes Streptomyces a very important alternative as a host for the expression of heterologous proteins of interest.
Our group has a lot of experience in studying the production of hydrolytic enzymes involved in the degradation of lignocellulosic materials by these bacteria. One of the fruits of this work has been the identification of several strong promoters regulated by the carbon source present in the culture media that have been used in different vectors for the overexpression of different proteins of industrial interest in Streptomyces (Diaz et al. 2008, 2011, Aragon et al. 2013). This study has been complemented with the development of expression vectors for Streptomyces based on a positive selection by not requiring the addition of antibiotics for their maintenance. For this purpose we have used the YoeB / YefM toxin-antitoxin system from S. lividans What we have During the development of this expression system we have generated a strain in which we have deleted the entire YoeB / YefM system from the genome and in which we have recently described and which has been the first toxin-antitoxin system functionally described in Streptomyces (Sevillian et al. 2012).
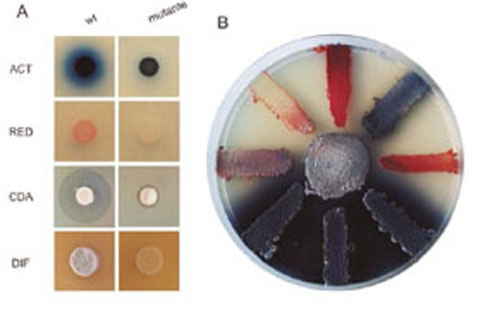
Figure 2. A. Effect of the mutation of a response regulator on the production of antibiotics by Streptomyces coelicolor and on their differentiation (DIF). Actinorhodine (ACT), undecylprodigiosin (RED), calcium-dependent antibiotic (CDA). B. Effect of different mutations in S. coelicolor on the production of antibiotics, the wild strain is the central one.
Representative bibliography
Rico S., Yepes, A., Rodríguez, H., Santamaría, J., Antoraz, S., Krause, E., Díaz, M and Santamaría, RI. (2014 a). Regulation of the AbrA1/A2 two-component system in Streptomyces coelicolor and the potential of its deletion strain as a heterologous host for antibiotic production. (PLOS One in press)
Rico S, Santamaría RI, Yepes A, Rodríguez H, Laing E, Bucca G, Smith CP, Díaz M. (2014b). Deciphering the regulon of the Streptomyces coelicolor AbrC3, a positive response regulator of antibiotic production. Appl. Environ. Microbiology 80(8): 2417-2428.
Rodríguez, H., Rico, S., Díaz, M. and Santamaría, RI. (2013). Two-component systems in Streptomyces: key regulators of antibiotic complex pathways. Microbial Cell Factories 12:127.
Sevillano, L., Díaz, M. and Santamaría, RI. (2013). Stable expression plasmids for Streptomyces based on a toxin-antitoxin system. Microbial Cell Factories 12:39
Díaz, M., Sevillano, L., Rico, S., Lombo, F., Braña, AF., Salas, JA., Méndez, C. and Santamaría, RI. (2013). High level of antibiotic production in a double polyphosphate kinase and phosphate-binding protein mutant of Streptomyces lividans. FEMS letters 342, 123-129.
Sevillano L., Díaz, M. and Santamaría, RI. (2012). Identification of the first functional toxin-antitoxin system in Streptomyces. PloS One 7(3): e32977.
Yepes, A., Rico, S., Rodríguez, A., Santamaría, RI. and Díaz, M. (2011). Novel two-components systems involved in antibiotic regulation in Streptomyces coelicolor. PloS One 6(5): e19980.
Pérez, J, Muñoz-Dorado J, Braña AF, Shimkets L J, Sevillano L and Santamaría RI. (2011). Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microbial Biotechnology 4:175-183
Esteban A, Díaz M, Yepes A and Santamaría RI. (2008). Expression of the pstS gene of Streptomyces lividans is regulated by the carbon source and is partially independent of the PhoP regulator. BMC Microbiology 8:201 doi:10.1186/1471-2180-8-201
Díaz, M., Ferreras, E., Moreno, R., Yepes, A., Berenguer, J, and Santamaría, RI., (2008). High-level overproduction of Thermus enzymes in Streptomyces lividans. Applied Microbiology and Biotechnology 79, 1001-1008
