
Group photo. Inigo Lasa Inigo Lasa
Like man, bacteria prefer to live in cities where the company of other bacteria facilitates the development of a matrix in which the bacteria live protected from changes in environmental conditions and in an environment where it is easier to accumulate degrading enzymes, retain water or communicate. The Bacterial Biofilms group began its journey in 1999 and develops its research activity at the Institute of Agrobiotechnology, a joint research center between the Public University of Navarra, the CSIC and the Government of Navarra. Its research focuses on molecular bases and genetic characteristics of the biofilm formation process of two pathogenic bacteria: Staphylococcus aureus and Salmonella enteritidis be. Enteritidis. The reason that led us to choose phylogenetically distant bacteria to carry out our studies was the conviction that this strategy would allow us to recognize those elements that are common to the biofilm formation process from those other elements that are specific to each bacterium. Staphylococcus aureus It is one of the pathogenic bacteria that causes major health problems in developed countries. This is due to (i) the enormous amount of virulence factors that it is capable of producing and that give it great versatility as a pathogen; (ii) the existence of multi-resistant strains to antibiotics (commonly known as MRSA); and (iii) their ability to adhere to medical implants and cause biofilm-mediated infections. This last characteristic was decisive when we decided to use it as a study model of the biofilm formation process. Throughout these years we have learned that the biofilm matrix of We have started to develop a system to co-culture different bacterial species that mimics the biofilms formed during a lung infection or wound healing process. These cultures are combined with lung epithelial cells to screen for antibacterial drugs. it may be of a protein nature (previously, there was the idea that the biofilm matrix is always polysaccharide in nature) and that a complex cascade of regulators whose connections are still not well understood intervenes in the regulation of exopolysaccharide synthesis (Toledo-Arana et al.it may be of a protein nature (previously, there was the idea that the biofilm matrix is always polysaccharide in nature) and that a complex cascade of regulators whose connections are still not well understood intervenes in the regulation of exopolysaccharide synthesis (Toledo-Arana et al.it may be of a protein nature (previously, there was the idea that the biofilm matrix is always polysaccharide in nature) and that a complex cascade of regulators whose connections are still not well understood intervenes in the regulation of exopolysaccharide synthesis (Toledo-Arana et al.it may be of a protein nature (previously, there was the idea that the biofilm matrix is always polysaccharide in nature) and that a complex cascade of regulators whose connections are still not well understood intervenes in the regulation of exopolysaccharide synthesis (Toledo-Arana et al.it may be of a protein nature (previously, there was the idea that the biofilm matrix is always polysaccharide in nature) and that a complex cascade of regulators whose connections are still not well understood intervenes in the regulation of exopolysaccharide synthesis (Toledo-Arana et al.it may be of a protein nature (previously, there was the idea that the biofilm matrix is always polysaccharide in nature) and that a complex cascade of regulators whose connections are still not well understood intervenes in the regulation of exopolysaccharide synthesis (Toledo-Aranaet al.it may be of a protein nature (previously, there was the idea that the biofilm matrix is always polysaccharide in nature) and that a complex cascade of regulators whose connections are still not well understood intervenes in the regulation of exopolysaccharide synthesis (Toledo-Arana et al.it may be of a protein nature (previously, there was the idea that the biofilm matrix is always polysaccharide in nature) and that a complex cascade of regulators whose connections are still not well understood intervenes in the regulation of exopolysaccharide synthesis (Toledo-Arana et al., 2014). We have also studied the two-component signal transduction systems that connect environmental stimuli with the matrix synthesis machinery. More recently, using transcriptomic analysis methodologies, we have discovered the existence of a global overlapping transcription process in Gram-positive bacteria. According to our results, which have been subsequently confirmed by other authors, the overlapping RNAs are digested by a specific RNase for double-stranded RNA, RNase III, to fragments of 19-21 nucleotides (Lase et al., 2014). We have also studied the two-component signal transduction systems that connect environmental stimuli with the matrix synthesis machinery. More recently, using transcriptomic analysis methodologies, we have discovered the existence of a global overlapping transcription process in Gram-positive bacteria. According to our results, which have been subsequently confirmed by other authors, the overlapping RNAs are digested by a specific RNase for double-stranded RNA, RNase III, to fragments of 19-21 nucleotides (Lase et al., 2014). This process of digestion of overlapping RNAs takes place throughout the genome and provides a very simple method to coordinate the expression of contiguous genes (Lasa et al., 2014). This process of digestion of overlapping RNAs takes place throughout the genome and provides a very simple method to coordinate the expression of contiguous genes (Lasa et al., 2014). This process of digestion of overlapping RNAs takes place throughout the genome and provides a very simple method to coordinate the expression of contiguous genes (Lasa We have started to develop a system to co-culture different bacterial species that mimics the biofilms formed during a lung infection or wound healing process. These cultures are combined with lung epithelial cells to screen for antibacterial drugs.. Studies aimed at investigating the function of the 3' UTR using as a study model the 3' UTR region of the icaR gene, a regulator of the synthesis of the main exopolysaccharide of the biofilm matrix, have shown that bacterial 3' UTRs can also contain regulatory elements of gene expression (Ruiz de Los Mozos et al.. Studies aimed at investigating the function of the 3' UTR using as a study model the 3' UTR region of the icaR gene, a regulator of the synthesis of the main exopolysaccharide of the biofilm matrix, have shown that bacterial 3' UTRs can also contain regulatory elements of gene expression (Ruiz de Los Mozos
. Studies aimed at investigating the function of the 3' UTR using as a study model the 3' UTR region of the icaR gene, a regulator of the synthesis of the main exopolysaccharide of the biofilm matrix, have shown that bacterial 3' UTRs can also contain regulatory elements of gene expression (Ruiz de Los Mozos Salmonella. Studies aimed at investigating the function of the 3' UTR using as a study model the 3' UTR region of the icaR gene, a regulator of the synthesis of the main exopolysaccharide of the biofilm matrix, have shown that bacterial 3' UTRs can also contain regulatory elements of gene expression (Ruiz de Los Mozos We have started to develop a system to co-culture different bacterial species that mimics the biofilms formed during a lung infection or wound healing process. These cultures are combined with lung epithelial cells to screen for antibacterial drugs. . Studies aimed at investigating the function of the 3' UTR using as a study model the 3' UTR region of the icaR gene, a regulator of the synthesis of the main exopolysaccharide of the biofilm matrix, have shown that bacterial 3' UTRs can also contain regulatory elements of gene expression (Ruiz de Los Mozos et al.. Studies aimed at investigating the function of the 3' UTR using as a study model the 3' UTR region of the icaR gene, a regulator of the synthesis of the main exopolysaccharide of the biofilm matrix, have shown that bacterial 3' UTRs can also contain regulatory elements of gene expression (Ruiz de Los Mozos et al., 2005). Subsequently, we focused on the signal transduction system mediated by c-di-GMP where we carried out an original approach that consisted in the elimination of the elements of this signal transduction pathway of this bacterium (García et al., 2005). Subsequently, we focused on the signal transduction system mediated by c-di-GMP where we carried out an original approach that consisted in the elimination of the elements of this signal transduction pathway of this bacterium (García et al., 2005). Subsequently, we focused on the signal transduction system mediated by c-di-GMP where we carried out an original approach that consisted in the elimination of the elements of this signal transduction pathway of this bacterium (Garcíaet al.. Studies aimed at investigating the function of the 3' UTR using as a study model the 3' UTR region of the icaR gene, a regulator of the synthesis of the main exopolysaccharide of the biofilm matrix, have shown that bacterial 3' UTRs can also contain regulatory elements of gene expression (Ruiz de Los Mozos
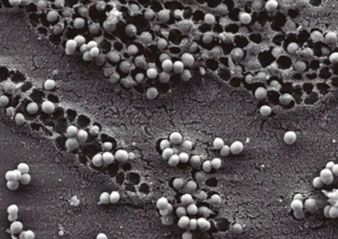
Figure 1. , 2005). Subsequently, we focused on the signal transduction system mediated by c-di-GMP where we carried out an original approach that consisted in the elimination of the elements of this signal transduction pathway of this bacterium (García
The use of this strain is allowing us to investigate the processes regulated by c-di-GMP in this bacterium and find out the specificity mechanisms that allow the c-di-GMP produced in response to a certain stimulus to specifically influence its targets, without interfering with the targets activated by other environmental stimuli. Although the main motivation of our research is to answer basic questions of when, how, and why bacteria form biofilms, we always try to identify biotechnological applications in our results. As a result of this vocation, the group has two international patents in operation and several members of the group have set up a technology-based company dedicated to the design and construction of genetically modified microorganisms "a la carte”. Our group maintains collaborations with many groups for the development of its research. We maintain a long and close collaboration with the group of Dr. José R. Penadés, previously in Segorbe, but who has recently moved to the University of Glasgow. We also maintain collaborations for the characterization of biofilm matrix proteins with the group of Dr. Jean Marc Ghigo (Pasteur Institute), the group of Dr. P. Romby (University of Strasbourg), Dr. Cecilia Arraiano (University Lisbon) and Dr. F. Vandenesh (University of Lyon) for the analysis of the overlapping transcription process. In the virulence and genetic aspects of Salmonella, we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville).
Representative bibliography
, we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). , we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). Salmonella , we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). Mol Microbiol , we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville)., we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville).
, we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). et al., we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). Staphylococcus aureus , we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). Infect Immun , we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville)., we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville).
, we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). , we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). PNAS, we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville)., we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville).
, we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville). (2012). An effort to make sense of antisense transcription in bacteria. (2012). An effort to make sense of antisense transcription in bacteria. Assay validation(2012). An effort to make sense of antisense transcription in bacteria.
(2012). An effort to make sense of antisense transcription in bacteria. et al. (2012). An effort to make sense of antisense transcription in bacteria. Proc Natl Acad Sci USA (2012). An effort to make sense of antisense transcription in bacteria.(2012). An effort to make sense of antisense transcription in bacteria.
(2012). An effort to make sense of antisense transcription in bacteria. (2012). An effort to make sense of antisense transcription in bacteria. Salmonella enterica (2012). An effort to make sense of antisense transcription in bacteria. Mol Microbiol (2012). An effort to make sense of antisense transcription in bacteria.(2012). An effort to make sense of antisense transcription in bacteria.
(2012). An effort to make sense of antisense transcription in bacteria. (2012). An effort to make sense of antisense transcription in bacteria. Escherichia coli. Proc Natl Acad Sci USA , we have always counted on the advice and help of the groups of Dr. F. García del Portillo (CNB, Madrid) and Dr. J. Casadesus (University of Seville).(2012). An effort to make sense of antisense transcription in bacteria.
Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., et al.Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., Staphylococcus aureus. J Bacteriol 191Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E.,
Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., et al.Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., Assay validationMerino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E.,
Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., 11Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E.,
Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., et al.(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci USA (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in
(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella enteritidis (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Mol Microbiol (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in
(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Staphylococcus aureus (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in ica(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in J Bacteriol (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in
(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in et al.(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Enterococcus faecalis (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Appl Environ Microbiol (2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in(2009). Genetic reductionist approac hfor dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in
Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. Staphylococcus aureus Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. 8Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I.
Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. Staphylococcus aureus. Mol Microbiol Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I.Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I.
Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. Staphylococcus aureus Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. J Bacteriol Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I.Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I.
Valle, J., Latasa, C., Gil, C., Toledo-Arana, A., Solano, C., Penadés, J.R., and Lasa, I. et al.(2009). Relevant role of fibronectin-binding proteins in Staphylococcus aureus (2009). Relevant role of fibronectin-binding proteins in Infect Immun (2009). Relevant role of fibronectin-binding proteins in(2009). Relevant role of fibronectin-binding proteins in
(2009). Relevant role of fibronectin-binding proteins in et al. (2009). Relevant role of fibronectin-binding proteins in Salmonella (2009). Relevant role of fibronectin-binding proteins in J Bacteriol (2009). Relevant role of fibronectin-binding proteins in(2009). Relevant role of fibronectin-binding proteins in
