Group photo: Laura Álvarez, Sara B. Hernández, Akbar Espaillat, Felipe Cava, Oskar Forsmo, Carlos Terriente, Teresa del Peso-Santos, Eleonore Skärfstad, Emilie Nordström, and Alena Aliashkevich.
The Biology and Genetics of the Bacterial Wall group (http://www.mims.umu.se/groups/felipe-cava.html) is a young group that started at the CBMSO (CSIC-UAM) in Madrid in 2011, and just a year ago we moved to Umeå, Sweden. Our group is part of the Department of Molecular Biology at Umeå University, MIMS-EMBL (Laboratory of Molecular Infection Medicine Sweden) and UCMR (Umeå Center for Microbial Research). MIMS and UCMR provide an interdisciplinary environment equipped with cutting-edge technologies in microscopy, metabolomics, NMR and advanced bioinformatic analysis.
Our main interest is the study of one of the main "Achilles' heels" of bacteria: the cell wall. We study how bacteria modulate their wall to adapt to environmental changes and to the different stages of the infection process. For this we make use of a wide variety of advanced analytical techniques, correlation microscopy and image analysis, combined with molecular genetics, biochemistry, bioinformatics and molecular biology. Our findings will allow us to provide answers to fundamental aspects of bacterial physiology as well as to develop new strategies to combat emerging infectious diseases.
We are looking for enthusiastic people who want to join our group as a postdoc. We encourage you to send us your CV and a cover letter.
Our projects
1- Biology of D-amino acids
In previous works we have described the role of D-amino acids as modulators of cell wall metabolism and their function as signaling/communication molecules (17). Although we initially detected non-canonical D-amino acids (NCDAA) in cultures of Vibrio cholerae, the pathogen that causes cholera, in fact, a wide variety of bacteria not necessarily taxonomically related can release high concentrations of NCDAA into the extracellular medium. Its accumulation also coincides with the transition to the stationary phase and negatively regulates the synthesis of the bacterial wall, so NCDAAs could be involved in coordinating the metabolism of the cell wall and the cytoplasm when nutrients are scarce. Since NCDAAs seem to be a common trait of various bacteria and we have an extensive collection (more than 1000 different species), we are currently investigating their role in various bacterial models and in different physiological processes such as sporulation, biofilm formation and the production of secondary metabolites.
We have also characterized the NCDAA incorporation pathways (16), which has a great impact on polymicrobial communities, since bacteria incapable of producing these molecules are nevertheless capable of incorporating them into their cell walls. We have recently carried out a biochemical and functional characterization of an entire family of broad-spectrum racemases, the multispecific enzymes responsible for NCDAA production in bacteria (8).
2- New models of cell wall growth in bacteria
Based on the ability of bacteria to incorporate various types of D-amino acids, we have designed strategies for monitoring cell wall growth and dynamics. in vivo by utilizing fluorescent D-amino acids and bioorthogonal chemistry (12, 13). The universality of this method has revolutionized the way in which cell wall dynamics and growth studies are performed today (PLoS Biol. 2013 Dec 11(12):e1001728; Nature 2013 Dec 11, NatCommun 2013 4:2856). Using advanced techniques such as correlation microscopy, we carry out a spatio-temporal follow-up of peptidoglycan remodeling dynamics in living cells.
3- Characterization of murein diversity in the Bacteria kingdom
Recently, we have developed online UPLC-MS technology for peptidoglycan high-through-put analysis that will allow us to analyze thousands of microbes under various conditions. This is a very ambitious research program that aims to discover and exploit the variability, both diversity and plasticity, of the bacterial cell wall. This research is critical to understanding cell wall biology in nature, the relationship of bacteria to other organisms, and their ability to adapt to environmental challenges. With the data obtained we are creating the first database of the bacterial cell wall: the MUREINOMA. This bacterial wall encyclopedia will contribute to the discovery of new metabolic and regulatory pathways of great interest in the development of new, more specific antimicrobial therapies.
4- Search for new modulating metabolites of the bacterial wall.
Another of our lines of research consists in studying the impact of environmental metabolites on the cell wall by using both libraries of chemical compounds and more complex biological extracts (marine extracts produced by Actinobacteria from the Arctic).
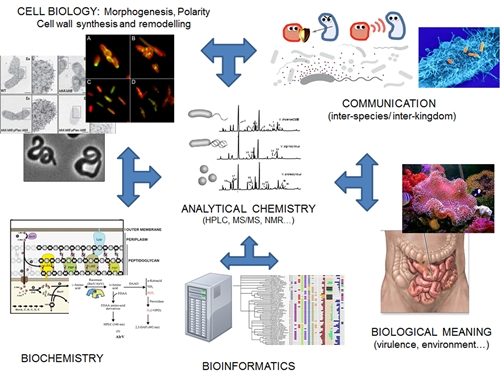
Figure 1. Diagram of the activities of the Bacterial Wall Biology and Genetics group. In our laboratory we carry out a multidisciplinary effort to understand the adaptive / physiological value that control over the composition and structure of its wall has for bacteria.
5- Development and dispersal of biofilm of Vibrio cholerae. The production of extracellular matrix is a common characteristic of multicellular communities, although there is great diversity in their formation. Recent studies suggest that some signaling molecules play an important role in the development of the biofilm. However, the molecular mechanisms responsible for the connection between chemical communication and biofilm architecture in polymicrobial communities remains a mystery.Our laboratory uses RNA seq technology to decipher molecular mechanisms by which certain environmental stimuli govern the development of the biofilm of Vibrio cholerae. Researchers of great international prestige work on the campus in the areas of molecular microbiology and infection biology. One of the main goals of our group is to train the next generation of scientists by creating a stimulating environment that encourages learning through collaboration.
Representative bibliography
Moll A, Dörr T, Alvarez L, Chao MC, Davis BM, Cava F, and Waldor MK. (2014). Cell separation in Vibrio cholerae is mediated by a single amidase whose action is modulated by two non-redundant activators. J Bacteriol. In press
Alvarez L, Espaillat A, Hermoso J A, de Pedro M A and Cava F. (2014). Peptidoglycan Remodeling by the Coordinated Action of Multispecific Enzymes. Microb Drug Resist.20(3):190-8
Hernández SB, Cava F, Pucciarelli MG, García-Del Portillo F, by Pedro MA, Casadesús J. (2014). Bile-induced peptidoglycan remodeling in Salmonella enterica. Environ Microbiol. doi: 10.1111/1462-2920.12491
Cava F and Pedro MA. (2014). Peptidoglycan plasticity in bacteria: emerging variability of the mureinsacculus and their associated biological functions. CurrOpinMicrobiol; 18C:46-53.
Dörr T, Lam H, Alvarez L, Cava F, Davis BM and Waldor MK. (2014). A Novel Peptidoglycan Binding Protein Crucial for PBP1A-Mediated Cell Wall Biogenesis in Vibrio cholerae. PLoSGenet. Jun 19;10(6):e1004433.
Dörr T, Möll A, Chao MC, Cava F, Lam H, Davis BM and Waldor MK. (2014). Differential requirement for PBP1a and PBP1b in the in vivo and in vitro fitness of Vibrio cholerae. InfectImmun. 82(5):2115-24.
Desmarais SM, Cava F, De Pedro MA and Huang KC. (2014). Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J Vis Exp. (83). doi: 10.3791/51183.
Espaillat A, Carrasco-Lopez C, Bernardo-Garcia N, Pietrosemolli N, Otero L H, Alvarez L, de Pedro M A, Pazos F, Davis B M, Waldor M K, Hermoso J A and Cava F. (2014). Structural basis for the broad specificity of a new family of amino acid racemases. Acta Crystallogr D BiolCrystallogr, 70:79-90
Cava F, Kuru E, Brun YV and de Pedro MA. (2013). Modes of cell wall growth differentiation in rod-shaped bacteria. CurrOpinMicrobiol. 16(6):731-7
Dorr T, Cava F, Lam H, Davis BM and Waldor MK (2013). Substrate specificity of an elongation-specific peptidoglycan endopeptidase and its implications for cell wall architecture and growth of Vibrio cholerae. MolMicrobiol, Jul 9, doi: 10.1111/mmi.12323.
Desmarais SM, De Pedro MA, Cava F and Huang KC (2013). Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. MolMicrobiol 89 (1): 1-13.
Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F y Bertozzi CR (2013). (D)-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS ChemBiol 8 (3): 500-505.
Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV y VanNieuwenhze MS (2012). In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids.AngewChemInt Ed Engl 51 (50): 12519-12523.
Horcajo P, by Pedro MA and Cava F (2012). Peptidoglycan plasticity in bacteria: stress-induced peptidoglycan editing by noncanonical D-amino acids. Microb Drug Resist 18 (3): 306-313.
Cava F, Lam H, by Pedro MA and Waldor MK (2011). Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell Mol Life Sci 68 (5): 817-831.
Cava F, by Pedro MA, Lam H, Davis BM and Waldor MK (2011). Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J 30 (16): 3442-3453.
Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA and Waldor MK (2009). D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325 (5947): 1552-1555.

