
Group photo. From left to right: Antonio Ventosa, Maribel Reina, Blanca Vera, Cristina Sánchez-Porro, Ana Durán, Rafael R. de la Haba, Cristina Galisteo
Our research group has been carrying out metagenomic studies in hypersaline environments, especially in coastal salt flats in our country, which are excellent models for studying extreme habitats, presenting a salinity gradient from that of seawater to salt saturation. These studies have made it possible to determine the structure and activity of the groups of archaea and bacteria that constitute the microbial diversity of these hypersaline systems, of which a high percentage of abundant microorganisms in these environments have not yet been isolated in pure culture. For this reason, an objective of our group is to carry out exhaustive studies focused on the isolation and characterization of these groups of archaea and halophilic bacteria from marine salt flats.
Among the new groups that we have recently isolated and characterized in detail are species of the genera Natronomonas, Halorientalis, Natrinema and Halorubrum, and very especially we must refer to the isolation of three new strains belonging to the genus Halonotius and four new strains that constitute a new genus, for which we propose the designation of Halosegnis, with two new species: Halosegnis long and Halosegnis rubeus. Genomic recruitment and ribosomal sequence comparison studies indicate that these new taxa constitute abundant groups in hypersaline environments, with a wide geographic distribution. Metabolic reconstructions based on the analysis of the genomes of the species of the genus Halonotius (Figure 1) demonstrated that they possess the genes for cobalamin biosynthesis (vitamin B12), a relevant result that would justify their wide geographic distribution and abundance in hypersaline environments, in which they could play an important role as a source of said vitamin. These studies are part of the Doctoral Thesis of Ana Durán Viseras, which will be presented recently.
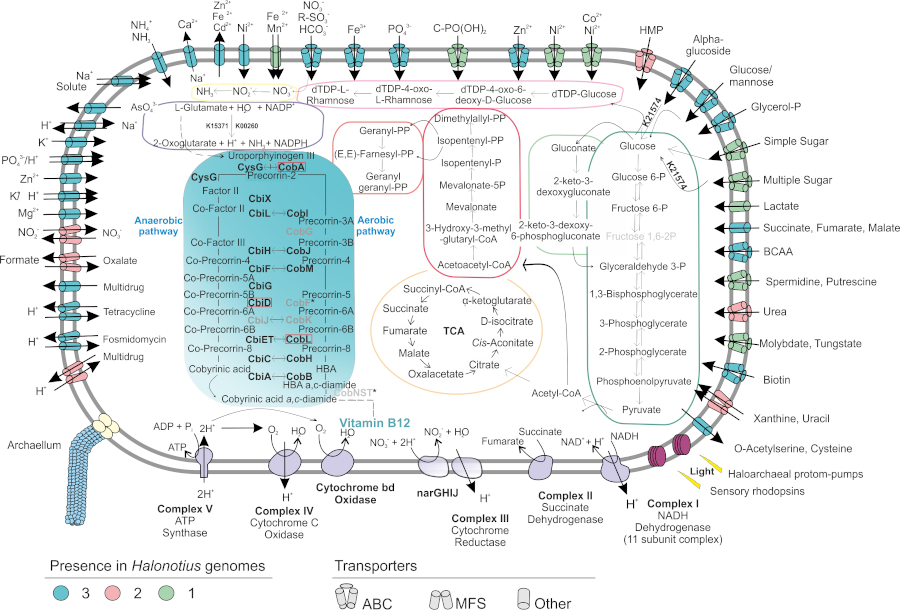
Figure 1. Metabolic reconstruction of haloarchaeas of the genus Halonotius (taken from Durán-Viseras et al., Front. Microbiol. 10: 1928, 2019).
Phylogenomic and functional diversity of prokaryotes in hypersaline soils
A research project that we are currently developing has as its objectives the determination of the structure and activity of the prokaryotic community of hypersaline soils, as well as the comparison of said terrestrial environments with the aquatic hypersaline environments that we have previously studied and of which there is much greater information. This study has been the subject of a Doctoral Thesis, carried out by Blanca Vera Gargallo (2018), in hypersaline soils located in the Marismas del Odiel, Huelva, through the use of metagenomic, metagenomic and stable isotope labeling (SIP) techniques. The hypersaline soils studied showed greater diversity, both taxonomic and functional, with respect to the previously studied aquatic environments, with the majority being groups belonging to Euryarchaeota, Bacteroidetes, Balneolaeota and Rhodothermaeota, although a high proportion of metagenomic sequences were affiliated with taxa not cultivated to date, especially belonging to the domain Bacteria. The SIP studies indicate that the archaea community of the saline soils studied is better adapted than the bacterial community present in these extreme environments (Vera-Gargallo & Sucker, 2018; Vera-Gargallo et al., 2019).
Gender Salinivibrio: MLSA and comparative genomics
The gender Salinivibrio includes moderate halophilic bacteria, normally found in hypersaline environments (saline, saline lakes), as well as in brines and salted foods. This genus includes a very small number of species and our research group has been carrying out studies on their taxonomic status, as well as the isolation of new strains of Salinivibrio that could constitute new species of this genus. For this purpose, and as part of the Doctoral Thesis study by Clara López Hermoso (2017), we isolated 70 new strains from saline environments in different locations. Due to the difficulty in differentiating these strains at the species level by comparing 16S rRNA sequences, we established an alternative methodology, based on MultiLocus Sequence Analysis, MLSA, by comparing housekeeping genes. gyrB, recA, rpoA and rpoB. This study was validated by performing alternative studies based on conventional DNA-DNA hybridization techniques, traditionally used in the delineation of species in prokaryotes (López-Hermoso et al., 2017). More recently, we have obtained the "draft" genomes of 36 strains of Salinivibrio, including the type strains of all described species and subspecies. On the other hand, by not having any closed genome of representatives of Salinibibrio, we have proceeded to complete genome sequencing of the type strain of Salinivibrio kushneri, a species recently described by our research group. Like other members of the family Vibrionaceae, S. kushneri it has two circular chromosomes with sizes of 2.84 Mb and 0.60 Mb, respectively; on the other hand, it has a high number (9) of ribosomal operons, typical of bacteria with rapid growth rates in laboratory media. Analysis of the genomes of strains of Salinivibrio revealed a high homogeneity and degree of synteny among the representatives of this genus. The phylogenomic study has allowed to know in detail the structure of the genus, being constituted by six species (S. costicola, S. siamensis, S. sharmensis, S. proteolyticus, S. kushneri and S. socompensis) and an additional species, which must be described in the future (from the Bean et al., 2019).
Selected publications (last 5 years)
Ventosa A, de la Haba RR, Sánchez-Porro, C, Papke, RT (2015). Microbial diversity of hypersaline environments: a metagenomic approach. Curr. Op. Microbiol. 25: 80-87.
Amoozegar MA, Siroosi M, Atashgahi S, Smidt H, Ventosa A. (2017). Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology 163: 623-645.
López-Hermoso C, de la Haba RR, Sánchez-Porro C, Papke RT, Ventosa A (2017). Assessment of Multilocus Sequence Analysis as a valuable tool for the classification of the genus Salinivibrio. Front. Microbiol. 8: 1107.
Oren A, Ventosa A, Kamekura M (2017). Class Halobacteria. Bergey’s Manual of Systematics of Archaea and Bacteria. W. B. Whitman (ed.), pp. 1-5. Whiley & Sons, New Jersey.
León MJ, Hoffmann T, Sánchez-Porro C, Heider J, Ventosa A, Bremer E (2018). Compatible solute synthesis and import by the moderate halophile Spiribacter salinus: physiology and genomics. Front. Microbiol. 9: 108.
Vera-Gargallo B, Ventosa, A (2018). Metagenomic insights into the phylogenetic and metabolic diversity of the prokaryotic community dwelling in hypersaline soils from the Odiel saltmarshes (SW Spain). Genes 9: 152.
de la Haba RR, Corral P, Sánchez-Porro C, Infante-Domínguez C, Makkay A, Amoozegar MA, Ventosa A, Papke RT (2018). Genotypic and lipid analyses of strains from the archaeal genus Halorubrum reveal insights into their taxonomy, divergence and population structure. Front. Microbiol. 9: 512.
Vera-Gargallo B, Chowdhury TR, Brown J, Fansler SJ, Durán-Viseras A, Sánchez-Porro C, Bailey VL, Jansson JK, Ventosa A (2019). Spatial distribution of prokaryotic communities in hypersaline soils. Sci. Rep. 9: 1769.
Windy A, de la Haba RR, Sánchez-Porro C (2019). Genus Haloarcula. Bergey’s Manual of Systematics of Archaea and Bacteria. W. B. Whitman (ed.), pp. 1-12. Whiley & Sons, New Jersey.
Amoozegar MA, Safarpour A, Noghabi KA, Bakhtiary T, Ventosa A. (2019). Halophiles and their vast potential in biofuel production. Front. Microbiol. 10: 1895.
Durán-Viseras A, Andrei A-S, Ghai R, Sánchez-Porro C, Ventosa A (2019). New Halonotius species provide genomics-based insights into cobalamin synthesis in haloarchaea. Front. Microbiol. 10: 1928.
de la Haba RR, López-Hermoso C, Sánchez Porro C, Konstantinidis K, Ventosa A (2019). Comparative genomics and phylogenomic analysis of the genus Salinivibrio. Front. Microbiol. 10: 2104.
