
Group photo. From left to right: Alba Vázquez Barca (doctoral student), Carlos Rodríguez Osorio (IP), Saqr Abushattal (doctoral student), Xosé Manuel Matanza Fente (doctoral student), Ana Vences Lorenzo (postdoctoral), Laura López Suárez (doctoral student).
The working group on Photobacterium damselae has more than 20 years of experience studying this important pathogen in marine fish aquaculture on a global scale. This species comprises two subspecies, damselae and Piscicides, which cause very different pathologies due to an interesting speciation process through which each subspecies has acquired mobile genetic elements carrying unique virulence factors.
P. damselae subsp. damselae It can be considered a general pathogen, which affects a wide variety of fish, as well as crustaceans and molluscs, and is an opportunistic pathogen for humans, and can cause fatal septicemia. Highly virulent strains contain a plasmid, pPHDD1, that encodes two potent cytotoxins, damselysin and phobalysin P, while a third hemolysin, phobalysin C, is chromosomally encoded. Damselysin acts synergistically with the two phobalysins to cause extensive cell damage, and our recent transcriptomics and proteomics studies demonstrate that the genes for these toxins are among the most highly expressed by this bacterium, the toxins are produced in large numbers, and are secreted by the type II secretion system. Strains that lack the pPHDD1 plasmid are less virulent in laboratory tests, but curiously, they are isolated with great frequency from outbreaks in fish farms, and in our group we have identified a new chromosomal phospholipase that contributes to virulence and cytotoxicity in these strains. .
This pathogen shows a high genetic diversity, and our recent studies show that genotypes of P. damselae subsp. damselae very diverse. Thus, strains with and without plasmids are isolated from the same batch of diseased fish, and the great diversity that exists in the genes that encode the polysaccharides of the cell envelope is of special relevance. This high genetic diversity led us to search for a weak point that was ubiquitous in the populations of this pathogen. Thus, we have recently discovered a two-component regulatory system that we have called RstAB, whose mutation practically cancels virulence, causes a drastic decrease in the expression of cytotoxin genes, and has negative consequences on the transcription of other virulence factors. additions, as well as in the cellular morphology. This system is present in all isolates of this subspecies and is a promising starting point for designing control strategies for this pathogen, as well as an interesting model for studying virulence regulation. At the moment we do not know the stimuli that trigger the activation of the RstAB system, although we have determined that a drop in the salinity of the medium stimulates the production of cytotoxins and other virulence factors.
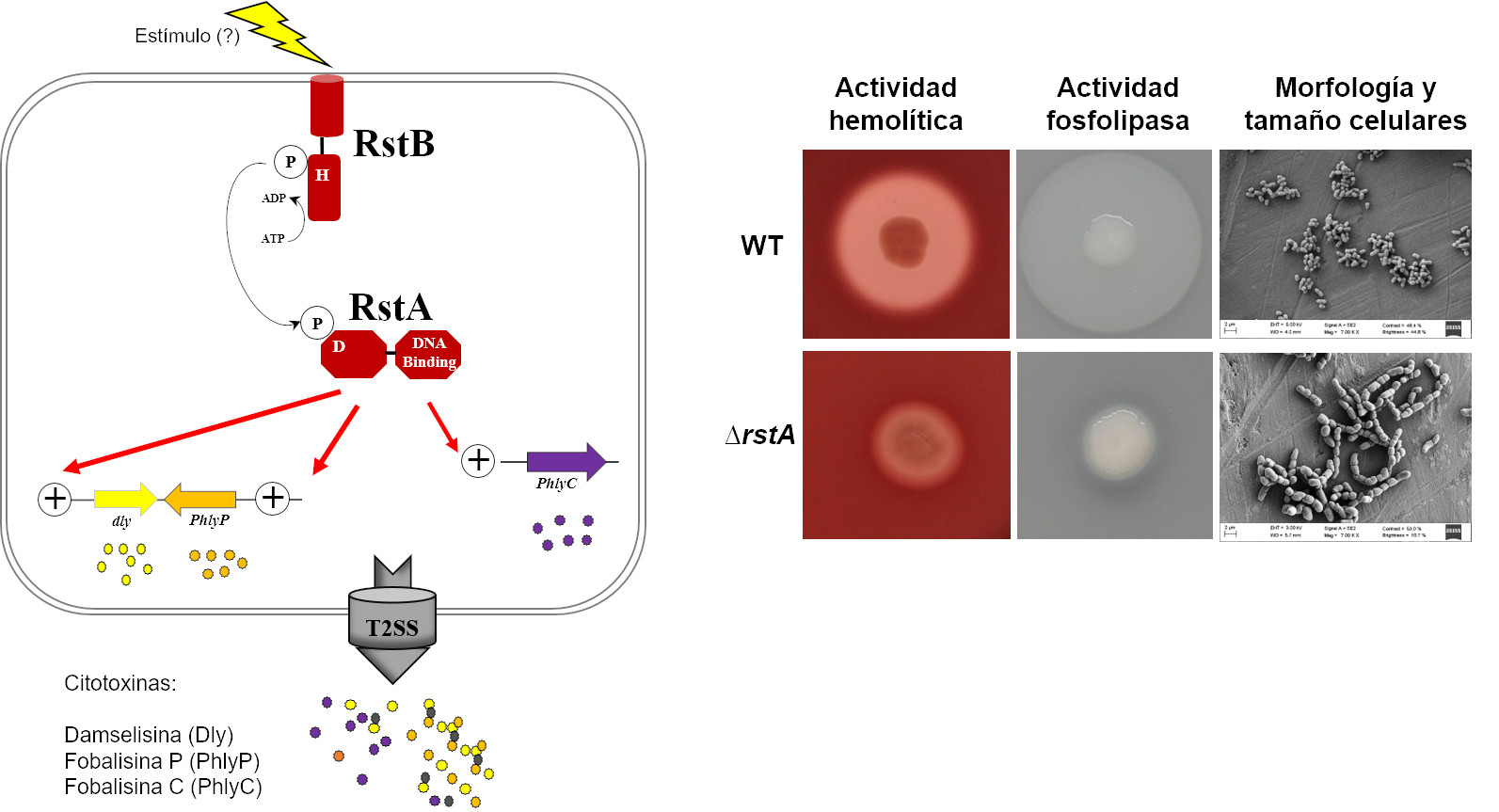
Figure 1. The two-component regulatory system RstAB regulates the three main cytotoxins of the pathogen P. damselae subsp. damselae, which are secreted by the type 2 secretion system (T2SS) and cause severe damage to host cells. In the right panel, it can be seen that the mutation of this system practically cancels the production of cytotoxins (with hemolytic and phospholipase activities), and greatly alters cell size, as well as the separation of daughter cells after division.
On the other hand, P. damselae subsp. Piscicides It is an exclusive pathogen of fish, in which it causes photobacteriosis or pseudotuberculosis, a disease of special importance in the Mediterranean area due to the great economic losses it causes in sea bream and sea bass farming. A complex process of accumulation of insertion elements has taken place in the genomes of the strains of this subspecies. As a consequence, in the genomes of subsp. Piscicides A massive reduction in gene functions has taken place, coupled with an evolution towards a way of life closely associated with the fish host. The best studied virulence factor in this subspecies is the toxin AIP56, which induces apoptosis in fish macrophages and neutrophils, and is secreted at high levels through the type II secretion system. This toxin is encoded on a small plasmid (pPHDP10) of only 9 kilobases. In addition, many strains of this subspecies contain a plasmid, pPHDP70, which includes in its structure a pathogenicity island with the genes for the synthesis and use of the siderophore piscibactin. Very recently, in our working group we have contributed to the first evidence of the existence of a type III secretion system in this subspecies. Far from being an anecdotal system in this pathogen, our recent data reveal a very extensive presence of this secretion system in subsp. Piscicides, which has opened new horizons in the study of the pathobiology of this subspecies.
Our current studies are aimed at applying the knowledge acquired in the more than 20 years of study of these two pathogens, to offer new strategies for diagnosis, prevention and control of diseases caused by both subsp. damselae as per subsp. Piscicides.
Recent group posts
Terceti MS, Vences A, Matanza XM, Barca AV, Noia M, Lisboa J, Dos Santos NMS, Do Vale A and Osorio CR. (2019). The RstAB system impacts virulence, motility, cell morphology, penicillin tolerance and production of Type II secretion system-dependent factors in the fish and human pathogen Photobacterium damselae subsp. damselae. Front Microbiol, 10:897.
Abushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR. Abushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR. Photobacterium damselae subsp. Piscicides Abushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR.
Abushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR. Abushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR. Photobacterium damselaeAbushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR. InAbushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR. & Abushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR.
Abushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR. Abushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR. Photobacterium damselae subsp. damselaeAbushattal S, Vences A, Dos Santos NMS, Do Vale A and Osorio CR.
Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Photobacterium damselae subsp. damselae Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR.
Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Photobacterium damselae subsp. Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR.
Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Photobacterium damselae subsp. damselae Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR.
Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR.. Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Terceti MS, Vences A, Matanza XM, Dalsgaard I, Pedersen K, and Osorio CR. Photobacterium damselae subsp. damselae major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582.
major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582. major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582. Photobacterium damselae subsp. damselaemajor virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582.. major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582.
major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582. major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582. Photobacterium damselae subsp. damselae major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582.major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582. major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582.major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582. major virulence factors Damselysin, Phobalysin P and Phobalysin C. Front Microbiol, 8:582.
