
Group photo. Dolores Lucía Guzmán-Herrador, Sara Samperio, Carlos Andrés Parra, Matxalen Llosa
Dolores Lucía Guzmán-Herrador, Sara Samperio, Carlos Andrés Parra, Matxalen Llosa
Our scientific interests focus on bacterial communication mediated by Type IV Secretion Systems (T4SS). These macromolecular complexes, like other families of secretion systems, are capable of translocating specific substrates across the membranes of gram-negative bacteria. What differentiates them from other systems and endows them with greater interest is their plasticity, being capable of secreting proteins as well as DNA and nucleoprotein complexes, both to the extracellular medium and to another recipient cell, whether prokaryotic or eukaryotic. This translates into an enormous biological versatility: the T4SS are part of the conjugative machinery to mediate horizontal gene transfer between bacteria, they are involved in the secretion of virulence factors to animal cells, they also play a role in symbiotic relationships between bacteria and animal cells. plants, inject toxins into competing bacteria, and are capable of secreting-importing DNA from the extracellular medium. These characteristics make them an interesting object of study from a biological, biotechnological and biomedical point of view. Based on the molecular knowledge we have of these processes, our group also intends to use these systems to develop tools for the introduction and site-specific integration of DNA in human cells.
A part of our work focuses on the comparative study of T4SS belonging to conjugative systems and pathogenic bacteria. Our study models are the T4SS of the conjugative plasmid R388, and the T4SS VirB/D4 of A part of our work focuses on the comparative study of T4SS belonging to conjugative systems and pathogenic bacteria. Our study models are the T4SS of the conjugative plasmid R388, and the T4SS VirB/D4 ofA part of our work focuses on the comparative study of T4SS belonging to conjugative systems and pathogenic bacteria. Our study models are the T4SS of the conjugative plasmid R388, and the T4SS VirB/D4 of
A very significant result of the group has been to show that substrates can be exchanged between T4SS involved in conjugation and virulence. Thus, we have shown that the nucleoprotein complexes that are transferred through the T4SS during bacterial conjugation, consisting of the conjugative relaxase covalently bound to the transferred DNA strand, can be recognized and translocated by the T4SS involved in the virulence of pathogens. humans A very significant result of the group has been to show that substrates can be exchanged between T4SS involved in conjugation and virulence. Thus, we have shown that the nucleoprotein complexes that are transferred through the T4SS during bacterial conjugation, consisting of the conjugative relaxase covalently bound to the transferred DNA strand, can be recognized and translocated by the T4SS involved in the virulence of pathogens. humans O A very significant result of the group has been to show that substrates can be exchanged between T4SS involved in conjugation and virulence. Thus, we have shown that the nucleoprotein complexes that are transferred through the T4SS during bacterial conjugation, consisting of the conjugative relaxase covalently bound to the transferred DNA strand, can be recognized and translocated by the T4SS involved in the virulence of pathogens. humans A very significant result of the group has been to show that substrates can be exchanged between T4SS involved in conjugation and virulence. Thus, we have shown that the nucleoprotein complexes that are transferred through the T4SS during bacterial conjugation, consisting of the conjugative relaxase covalently bound to the transferred DNA strand, can be recognized and translocated by the T4SS involved in the virulence of pathogens. humans
This result could be observed with little or no manipulation of natural systems, arguing that it possibly reflects a natural phenomenon, and horizontal DNA transfer between pathogens and their host cells, mediated by T4SS, may occur. Our current goal is to demonstrate whether this gene transfer does indeed occur in nature, and to discover the biological role it may play, either by contributing to the virulence of the pathogen encoded by T4SS, or by contributing to possible symbiotic relationships of the microbiome with its host.
The applied aspect of this line consists of manipulating the T4SS of different pathogenic bacteria so that they secrete DNA molecules of interest in vivo directly to the human cells of choice, which would be different depending on the tropism of the pathogen of choice.
Another line of work focuses on the characterization of conjugative relaxases, the proteins that process and guide DNA to the recipient cell during conjugation. Our model is the relaxase of the conjugative system of R388, TrwC. This protein, in addition to its role in conjugation, has site-specific recombinase and integrase activity in the recipient bacteria, an activity that we have characterized in bacteria and are testing in human cells. One of our challenges is to elucidate what would be the biological role of this unexpected activity, which curiously is shared by some, but not all conjugative relaxases analyzed. Our data indicate that, although the protein initiates recombination/integration reactions with high sequence specificity for its specific target, the targeting in the recipient cell is more lax. This leads us to think that such a system could provide the conjugative plasmid with a colonization system for non-permissive hosts (i.e. bacteria to which the plasmid can conjugate, but where it could not replicate), promoting its integration into the recipient genome.
By fine-tuning the T4SS DNA transfer assay of human pathogens, we have also studied the activity of TrwC and other relaxases after being transferred into human cells, discovering that relaxase enhances integration into cells by two orders of magnitude. the human genome of DNA transferred from the bacterium. This integration is not site-specific, although we are developing different strategies to make it so. Although we are still in the proof-of-concept stage, the potential applications are enormous.
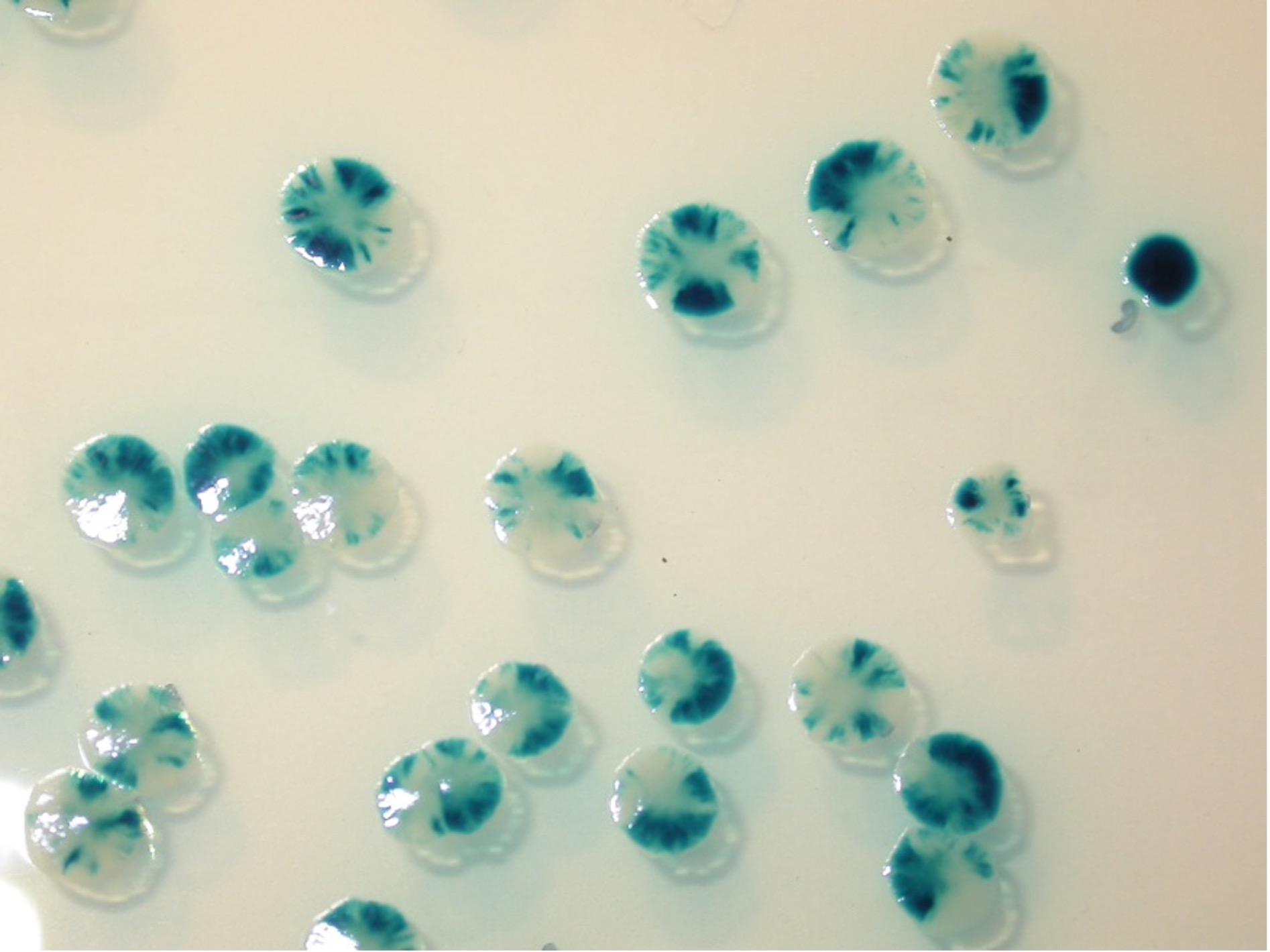
Figure. By fine-tuning the T4SS DNA transfer assay of human pathogens, we have also studied the activity of TrwC and other relaxases after being transferred into human cells, discovering that relaxase enhances integration into cells by two orders of magnitude. the human genome of DNA transferred from the bacterium. This integration is not site-specific, although we are developing different strategies to make it so. Although we are still in the proof-of-concept stage, the potential applications are enormous.
By fine-tuning the T4SS DNA transfer assay of human pathogens, we have also studied the activity of TrwC and other relaxases after being transferred into human cells, discovering that relaxase enhances integration into cells by two orders of magnitude. the human genome of DNA transferred from the bacterium. This integration is not site-specific, although we are developing different strategies to make it so. Although we are still in the proof-of-concept stage, the potential applications are enormous.
- Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M, Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,
- Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M, & Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,
Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M, - Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M, Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,
Front Microbiol. 2017 Aug 22; 8:1503. doi: 10.3389/fmicb.2017.01503 - Front Microbiol. 2017 Aug 22; 8:1503. doi: 10.3389/fmicb.2017.01503 Front Microbiol. 2017 Aug 22; 8:1503. doi: 10.3389/fmicb.2017.01503
Front Microbiol. 2017 Aug 22; 8:1503. doi: 10.3389/fmicb.2017.01503 - Front Microbiol. 2017 Aug 22; 8:1503. doi: 10.3389/fmicb.2017.01503 Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,.
Front Microbiol. 2017 Aug 22; 8:1503. doi: 10.3389/fmicb.2017.01503 - Front Microbiol. 2017 Aug 22; 8:1503. doi: 10.3389/fmicb.2017.01503 Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,.
Front Microbiol. 2017 Aug 22; 8:1503. doi: 10.3389/fmicb.2017.01503 - A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL, Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,.
A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL, - A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL, Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,.
A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL, - A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL,Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL,
A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL, - A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL, Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. Agúndez L, Zárate-Pérez F, Meier AF, Bardelli M,A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL,
A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL,
