
Group photo. From left to right: Sara Belén Hernández Piñero, Agustín Vioque Peña, Alicia M. Muro Pastor, Isidro Álvarez Escribano and Belén Suárez Murillo.
alicia@ibvf.csic.es2) by nitrogenase, an enzyme extremely sensitive to oxygen whose activity is incompatible with oxygenic photosynthesis. In complex cyanobacteria, like our model organism Nostoc ) by nitrogenase, an enzyme extremely sensitive to oxygen whose activity is incompatible with oxygenic photosynthesis. In complex cyanobacteria, like our model organism & Hess 2012). Heterocyst differentiation, which is ultimately controlled by NtcA, is also dependent on HetR, a specific regulator of cell differentiation. Our work focuses on the study of the participation of regulatory RNAs, both small RNAs (sRNAs) and antisense RNAs in the processes of adaptation to nitrogen deficiency stress and heterocyst differentiation.
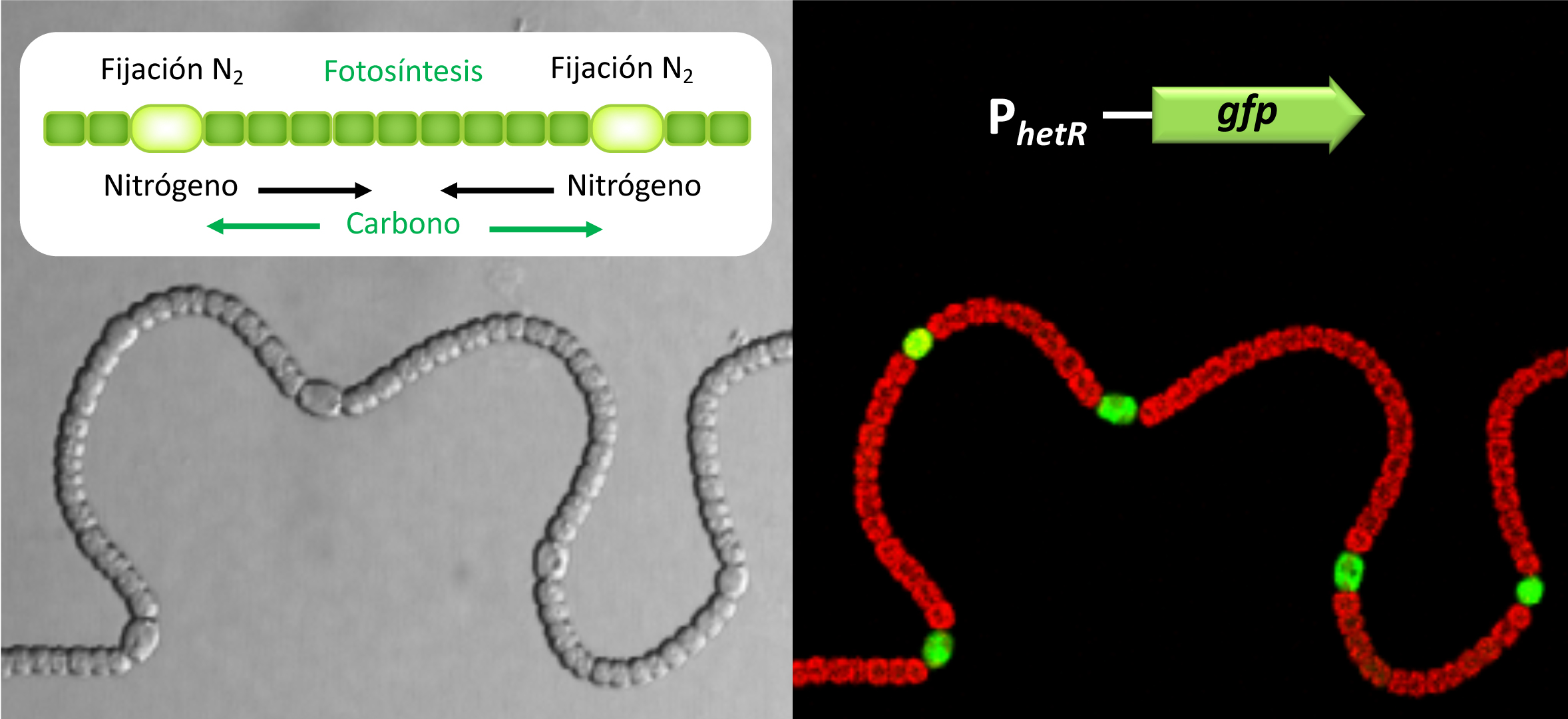
Figure 1. Hess 2012). Heterocyst differentiation, which is ultimately controlled by NtcA, is also dependent on HetR, a specific regulator of cell differentiation. Our work focuses on the study of the participation of regulatory RNAs, both small RNAs (sRNAs) and antisense RNAs in the processes of adaptation to nitrogen deficiency stress and heterocyst differentiation.2). Expression of the reporter protein GFP from the promoter of the differentiation regulator HetR illustrates the existence of transcriptional patterns unique to heterocysts. The red fluorescence corresponds to the photosynthetic pigments of the vegetative cells.
). Expression of the reporter protein GFP from the promoter of the differentiation regulator HetR illustrates the existence of transcriptional patterns unique to heterocysts. The red fluorescence corresponds to the photosynthetic pigments of the vegetative cells. hetR, which is not able to differentiate heterocysts, allowed us to classify transcriptional responses to nitrogen deficiency into two large regulons, the NtcA regulon, for global responses, and the HetR regulon, which includes responses specifically related to heterocyst differentiation (Mitschke et al, which is not able to differentiate heterocysts, allowed us to classify transcriptional responses to nitrogen deficiency into two large regulons, the NtcA regulon, for global responses, and the HetR regulon, which includes responses specifically related to heterocyst differentiation (Mitschke
, which is not able to differentiate heterocysts, allowed us to classify transcriptional responses to nitrogen deficiency into two large regulons, the NtcA regulon, for global responses, and the HetR regulon, which includes responses specifically related to heterocyst differentiation (Mitschke et al. 2016). This approach implements a phylogenetic conservation analysis suggesting that some of the predicted sRNAs would in fact be transcribed as functional RNAs. Thus, for example, NsrR1 (nitrogen stress-repressed RNA 1) is a 45-nucleotide sRNA, encoded in all the genomes of heterocyst-forming strains, whose expression is repressed by NtcA and which participates in the regulation of protein expression NblA, involved in the recycling of amino acids in conditions of nitrogen deficiency (Álvarez-Escribano et al. 2018). In this way, the regulation of NblA protein levels would be operated by a mixed circuit in which a transcriptional factor (NtcA) and an sRNA (NsrR1) participate. We have also analyzed NsiR4 (nitrogen stress-induced RNA4), which is transcribed from a promoter activated by NtcA and which in a unicellular cyanobacterium participates in the regulation of ammonium assimilation through the modulation of the translation of the IF7 protein, a glutamine synthetase inactivating factor (Klähn et al. 2015). The version of NsiR4 present in complex heterocyst-forming cyanobacteria modulates enzymatic activities involved in CO assimilation2 (Brenes-Álvarez et al., 2021).
We have also designed a global approach to the identification of heterocyst-specific transcripts, among which we have found both sRNAs and antisense RNAs that could play a regulatory role. From hybridization data of microarrays We have built a co-expression network and carried out a “clustering” analysis in which the different transcripts appear grouped based on common transcriptional patterns (Brenes-Álvarez et al. 2019). Among the transcripts transcriptionally grouped with those of genes involved in heterocyst differentiation, there appears, for example, NsiR1 (nitrogen stress-induced RNA1), a 60-nucleotide sRNA that can be considered a very early marker of differentiating cells (Muro-Pastor 2014) that we have shown that it affects the initial stages of heterocyst differentiation (Brenes-Álvarez et al., 2020, 2022). Likewise, among the transcripts that are expressed exclusively in heterocysts, several antisense transcripts appear, such as as_glpX. 2019). Among the transcripts transcriptionally grouped with those of genes involved in heterocyst differentiation appears, for example, NsiR1 (nitrogen stress-induced RNA1), a 60-nucleotide sRNA that can be considered a very early marker of cells in differentiation (Muro-Pastor 2014) and whose regulatory role we are currently analyzing. Likewise, among the transcripts that are expressed exclusively in heterocysts, several antisense transcripts appear, such as2 or the as_gltA, which is transcribed on the opposite strand of the gene that encodes citrate synthase. The transcription of these antisense RNAs specifically in heterocysts would contribute to the metabolic reprogramming that accompanies the differentiation of these specialized cells (Olmedo-Verd et al. 2019, Brenes-Alvarez et al., 2023).
In summary, in recent years we have contributed to defining mixed regulatory circuits that involve the transcriptional regulators NtcA and HetR and non-coding RNA molecules (both small RNAs and antisense RNAs) whose expression is under their control. Our work is carried out in collaboration with the laboratory led by Wolfgang R. Hess (Genetics and Experimental Bioinformatics, University of Freiburg, Germany) and is currently funded by the State Research Agency within the framework of the State Program for Knowledge Generation and Scientific and Technological Strengthening of the R&D&i System (PID2022-138128NB-I00).
https://www.ibvf.us-csic.es/grupos-investigacion/rnas-reguladores-de-cianobacterias/
Representative bibliography
Álvarez-Escribano I, Suárez-Murillo B, Brenes-Álvarez M, Vioque A, Muro-Pastor AM. (2024) Antisense RNA regulates glutamine synthetase in a heterocyst-forming cyanobacterium. Plant Physiol. 195:2911-2920.
Brenes-Álvarez, M., Vioque, A., Muro-Pastor, A. M. (2023) Nitrogen-regulated antisense transcription in the adaptation to nitrogen deficiency in Nostoc. PNAS Nexus, 2: pgad187.
Brenes-Álvarez, M., Vioque, A., Muro-Pastor, A. M. (2022) The heterocyst-specific small RNA NsiR1 regulates the commitment to differentiation in Nostoc. Microbiology Spectrum 10: e02274-21.
Song, K., Baumgartner, D., Hagemann, M., Muro-Pastor, AM, Maass, S., Becher, D., and Hess W.R. (2022) AtpΘ is an inhibitor of F0F1 ATP synthase to arrest ATP hydrolysis during low-energy conditions in cyanobacteria. Current Biology 32: 136-148.
In summary, in recent years we have contributed to defining mixed regulatory circuits involving the transcriptional regulators NtcA and HetR and non-coding RNA molecules whose expression is under their control. Our work is carried out in collaboration with the laboratory led by Wolfgang R. Hess (Genetics and Experimental Bioinformatics, University of Freiburg, Germany) and is currently funded by the State Research Agency within the framework of the State Program for the Generation of Knowledge and Scientific and Technological Strengthening of the R&D System (PID2019-105526GB-I00). (2021) A nitrogen stress inducible small RNA regulates CO2 (2021) A nitrogen stress inducible small RNA regulates CO Nostoc(2021) A nitrogen stress inducible small RNA regulates CO
(2021) A nitrogen stress inducible small RNA regulates CO (2021) A nitrogen stress inducible small RNA regulates CO Nostoc (2021) A nitrogen stress inducible small RNA regulates CO
(2021) A nitrogen stress inducible small RNA regulates CO (2020) NsiR1, a small RNA with multiple copies, modulates heterocyst differentiation in the cyanobacterium Nostoc sp. PCC 7120. Environmental Microbiology 22: 3325-3338.
(2021) A nitrogen stress inducible small RNA regulates CO (2021) A nitrogen stress inducible small RNA regulates CO Nostoc sp. PCC 7120. mBio 11: e02599-19.
sp. PCC 7120. mBio 11: e02599-19. sp. PCC 7120. mBio 11: e02599-19. Nostoc sp. PCC 7120. mBio 11: e02599-19.
sp. PCC 7120. mBio 11: e02599-19. sp. PCC 7120. mBio 11: e02599-19. Nostoc sp. PCC 7120. mBio 11: e02599-19.
(2019). CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol 16: 518-529. (2019). CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol 16: 518-529. (2019). CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol 16: 518-529. in Nostoc (2019). CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol 16: 518-529.
(2019). CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol 16: 518-529. (2019). CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol 16: 518-529.: (2019). CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol 16: 518-529.
(2019). CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol 16: 518-529. (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112: (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112
(2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112 (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112
(2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112 (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112
(2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112 (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112 Anabaena (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA 112
