
Group photo. Pedro Belda, Raúl Cabrera, Anny Camelo, Mariam Ferrer, Arantxa López, Áurea Simón and Alex Mira Pedro Belda, Raúl Cabrera, Anny Camelo, Mariam Ferrer, Arantxa López, Áurea Simón and Alex Mira
The Laboratory for the Study of the Human Microbiome is directed by Dr. Alex Mira Obrador, and was created in 2009, when it was integrated into the Genomics and Health Area of the FISABIO Foundation. The group's research focuses on characterizing the commensal and pathogenic human microbiota from a mixed approach of bioinformatics and experimental work in genomics and metagenomics. It is a multidisciplinary team that includes microbiologists, molecular biologists, dentists, pharmacists and bioinformaticians.
(Group photo). Dr Mira has been one of the founders of the National Bacterial Genomics Network. His scientific contribution to this field earned him the Jaime Ferrán Award from the Spanish Society of Microbiology in 2009. The group is made up of three predoctoral students (two of them in their final thesis phase), two postdoctoral researchers and a Master's student, and each year has 2-4 students in national and international internships. The training offered in the group is a mixed approach of experimental and computational techniques. The Laboratory holds weekly meetings for the management and discussion of scientific articles, and conducts an annual course on massive sequence analysis to study biodiversity.
Microbiome and disease diagnosis
We have used massive sequencing techniques to identify possible microorganisms associated with colorectal cancer. Biopsies of tumors and polyps showed a microbiota clearly different from that located in healthy areas of the colon (Mira-Pascual et al. 2014), which is very promising to use the microbial community as biomarkers of cancer before the appearance of clinical signs , which has recently been patented in collaboration with the company Entrechem SL.
Regarding dental caries, we have determined that the bacteria present in saliva are not good markers of this pathology. However, we have identified a series of metabolites that, when measured in this fluid, tell us with great precision if the person has cavities. This discovery has been patented as a caries risk diagnostic test, and by indicating the cause of the risk, it will allow personalized preventive treatment to be given to each patient.
Real-time biofilm formation
The group has developed a new in vitro biofilm model where the growth of monospecific and complex biofilms (such as dental plaque) can be monitored in real time. This biofilm model is being used to identify Quorum Sensing inhibitory molecules and to measure the growth of infection-causing bacteria on medical catheters and implants in real time, identifying not only the necessary inhibitory doses but also that some antibiotics actually stimulate the production of more biofilm. All this will be a huge economic savings in the health system, since the resistance to antibiotics of the bacteria that cause post-operative infections could be seen and an effective treatment could be decided in half the current time.
Milk contains microorganisms
The group has applied massive sequencing techniques to decipher the enormous diversity of bacteria contained in colostrum and breast milk (Cabrera-Rubio et al. 2012), work that may have enormous significance in the field of infant nutrition. We have found these bacteria in fecal samples from babies, so we believe that milk may be the solution that human evolution has found for a controlled transmission of microorganisms from one generation to the next. The role of the microbiota in breast milk is yet to be determined, and although we cannot rule out a metabolic role, we work with the hypothesis that these bacteria are essential for the proper development of the baby's immune system, affecting diseases such as asthma or allergies, which epidemiologically are found in greater proportions in children fed with formula milk.
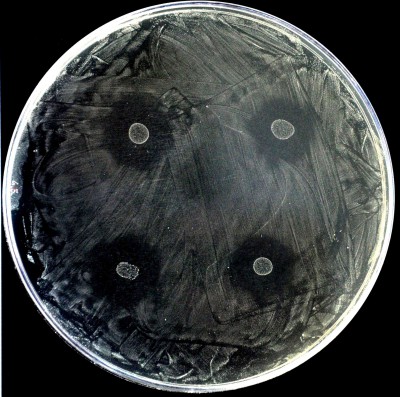
Figure. Tapestry culture of the caries-causing bacteria Streptococcus mutans, which is inhibited by the bacteriocins produced by four colonies of Streptococcus dentisani, currently developed as an anti-caries probiotic.
Other microbiomes
The lower respiratory tract, traditionally considered sterile, contain an enormous diversity of microorganisms, at least in patients with Chronic Obstructive Pulmonary Disease, and this bacterial population varies depending on the episodes of stability or crisis of the patient (Galiana et al. 2014). We have also determined that sputum, the sample of choice in most clinical studies because it is non-invasive, is not at all representative of the microbial diversity in the lung mucosa (Cabrera-Rubio 2012b). We have also collaborated with the IPLA-CSIC and the Hospital de Elche in studies of the stomach microbiota through pyrosequencing, showing that it enjoys enormous diversity, especially in the absence of Helicobacter pylori (Delgado et al. 2013), a bacterium in which we are also interested due to its possible involvement in autoimmune diseases.
Bacterial genomics
The most traditional work of the group was focused on microbial comparative and evolutionary genomics and, although it is no longer our main line of research, we continue to carry out work in this field. For example, in the concept of the bacterial pan-genome (Mira et al. 2010), in the identification of islands of pathogenesis through metagenomic recruitment (Belda-Ferre et al. 2011) or in a new method for the characterization of replication origins in prokaryotes. Using this approach, based on massive sequencing, we have identified four origins of replication in archaea. Pyrobaculum (pelvis et al. 2012).
Representative bibliography
Belda-Ferre P, Cabrera-Rubio R, Moya A, Mira A. (2011). Mining virulence genes using metagenomics. PLoS One 6(10):e24975.
Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simón-Soro A, Pignatelli M, Mira A. (2012). The oral metagenome in health and disease. ISME J. 6(1):46-56.
Benítez-Páez A, Belda-Ferre P, Simón-Soro A, Mira A. (2014). Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics. 15:311.
Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira_A. (2012a). The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J
Clin Nutr. 96(3):544-51.
Cabrera-Rubio R, Garcia-Núñez M, Setó L, Antó JM, Moya A, Monsó E, Mira A. (2012b). Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol.
50(11):3562-8.
Camelo-Castillo A, Benítez-Páez A, Belda-Ferre P, Cabrera-Rubio R, Mira A. (2014). Streptococcus dentisani sp. nov., a novel member of the mitis group. Int J Syst Evol Microbiol. 64(Pt 1):60-5.
Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. (2013). Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol. 65(3):763-72.
Galiana A, Aguirre E, Rodriguez JC, Mira A, Santibañez M, Candela I, Llavero J, Garcinuño P, López F, Ruiz M, Garcia-Pachon E, Royo G. (2014). Sputum microbiota in moderate versus severe patients with COPD. Eur Respir J. 43(6):1787-90.
Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F. Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F.
Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F. Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F.
Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F. Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F. Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F.Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F.
Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F. (2013). A tissue-dependent hypothesis of dental caries. Caries Res. 47(6):591-600.
Simón-Soro A, Guillen-Navarro M, and Mira A. (2014). Metatranscriptomics reveals overall active bacterial composition in caries lesions. J Oral Microbiol 6:25443.
