
agcampa@isciii.es From left to right: Alejandro Arce, José Manuel Borrero, Andony Flores, Camilo Vásquez, Cristina Civantos, Patricia Bernal, Javier de la Peña, Adrián Ruiz and María Olmo
Our group is made up of two main researchers and their teams with convergent lines of research: Patricia Bernal Guzman, y Jose Manuel Borrero de Acuña.
We study the molecular mechanisms of different secretion systems, especially the type VI Secretion System (T6SS) and the bacterial Extracellular Membrane Vesicles (EMV), which have been proposed as the type 0 secretion system (T0SS).
In recent years, the two research groups have combined their knowledge to carry out a new line of research in the study of blood vesicles. Pseudomonas. The interest of this new line lies in their characterization, the multiprotein complexes involved in their biogenesis, as well as their bioengineering to transport antimicrobial agents in the fight against pathogenic microorganisms. This line of basic science research has potential biotechnological applications in the field of sustainable agriculture and livestock, as well as biomedicine by studying molecular mechanisms of bacterial growth control.
The members of the group are detailed below in alphabetical order:
o Arce Rodríguez, Alejandro – Senior postdoctoral researcher
o Ayala García, Paula – Postdoctoral researcher
o Azogue Palma, Carlos – Hired project intern
o Bernal Guzmán, Patricia – Hired Ramón y Cajal
o Borrero de Acuña, José Manuel – Hired Emergia
o Civantos Jiménez, Cristina – Laboratory technician
o de la Peña Noya, Javier – Predoctoral fellow hired by project
o Herrero Gómez, Irene – FPU predoctoral fellow
o Moreno de Castro, Natalia – FPU predoctoral fellow
o Paredes Bermejo, María del Carmen – Laboratory technician
o Ruiz Camas, Adrián – Laboratory technician
Joint projects
- Title of the project: Engineering extracellular membrane vesicles from rhizospheric bacteria for the development of biopesticides and plant-growth promoting agents.
- Financing entity: Projects of Excellence 2021. General Secretariat of Universities, Research and Technology, Junta de Andalucía.
- Reference: ProyExcel_00450 Lead Entity: Sevilla University Period: December 2022-November 2025
- Subsidy amount: 140.937,14€ Responsible Researchers: Jose Manuel Borrero de Acuña & Dr. Patricia Bernal
- Title of the project: Development of biological pesticides based on membrane vesicles as a sustainable alternative to highly polluting chemical pesticides (BIOPESTOMV).
- Financing entity: Ministry of Science and Innovation, Strategic Projects Oriented to the Ecological Transition and the Digital Transition 2021, Government of Spain
- Reference: TED2021-130357B-I00 Lead Entity: Sevilla University Period: December 2022-November 2024
- Subsidy amount: 253.000,00€ Responsible Researchers: Dr. Patricia Bernal & Jose Manuel Borrero de Acuña
Joint Predoctoral Students
Javier de la Peña Noya – Characterization and bioengineering of the VEMs of Pseudomonas putida KT2440.
Joint publications
- Borrero de Acuña, J.M., & Bernal, P.*; (2021). Plant holobiont interactions mediated by the type VI secretion system and the membrane vesicles: promising tools for a greener
agriculture. Environmental Microbiology, 23(4), 1830-1836.
Dissecting the multiprotein complexes and molecular mechanisms sustaining bacterial membrane vesicles biogenesis: towards extracellular membrane vesicle cargo engineering
PI: Jose Manuel Borrero de Acuña
Research in our lab:
One of our major endeavours is to elucidate protein complex assembly dynamics involved in crucial biological processes for diverse bacteria. The majority of cellular functions in all living cells are conducted by proteins. Biochemical pathways, signal transduction cascades, and membrane-associated complexes of energy generation rely on the fine-tuned, directed and frequently transient protein interactions. We have widely investigated biochemical processes for relevant human pathogens (Legionella pneumophila, Clostridioides difficile and Pseudomonas aeruginosa) and environmentally relevant bacteria (Dinoroseobacter shibae and P. putida); (see publications: https://orcid.org/my-orcid?orcid=0000-0002-6409-8110). Dissecting the protein-protein interactions underlying such processes is crucial for basic research but it is also a cornerstone for boundless biomedicine and biotechnological applications. We also investigate the molecular processes governing organisms’ interactions in their natural niche. Our interest is drawn towards the elucidation of the biogenesis pathways and cell-to-cee communication mechanisms driven by extracellular membrane vesicles (EMVs). EMVs are nanoparticles involved in a broad range of biological processes including horizontal DNA transfer, decoy for phages and antibiotics, disposal of waste material and surface remodeling, nutrient scavenging, bacterial killing, host response immunomodulation and delivery of bioactive compounds and thus are essential for inter- intra-species and inter-kingdom communication. Recently, interactomic-driven research has led us to the identification of highly conserved proteins sustaining EMV formation across species. In our current projects we propose to exploit the full potential of EMVs for the benefit of bacteria-plant interactions. To this end, we employ interactomics techniques consisting of affinity purification coupled with mass spectrometry, SPOT-membrane arrays, immunofluorescence and immunogold labelling microscopy and others to identify further protein-protein interactions underlying the biogenesis of EMVs in different bacterial species present in the plant holobiont. We induce the expression of these proteins found in the interactomic studies to modulate vesicle formation rate, size and amount. We intend to tailor the protein and metabolic cargo of EMVs by encapsulating nodulation factors, plant-priming molecules, nitrogen nitrogen-fixing enzymes or phytopathogen inhibitors. Thereby, EMVs can be used as organism-free biopesticides for phytopathogen killing or as bioinoculants to enhance nodulation, nitrogen fixation and ultimately plant growth. Nonetheless, EMV research go beyond the applications related to sustainable agriculture since novel molecular tools arising from this work can be applied in multiple disciplines, ranging from biomedicine (development of vaccines) to bioremediation (design of EMV-based xenobiotic degraders).
Dissection of the multiprotein complexes and molecular mechanisms underlying bacterial membrane vesicle biogenesis: toward engineering extracellular membrane vesicle cargo.
Research lines in our laboratory:
Our group focuses broadly on elucidating the assembly dynamics of protein complexes involved in crucial biological processes for various bacteria. Most cellular functions in all living cells are performed by proteins. Biochemical pathways, signal transduction cascades, and membrane-associated energy generation complexes depend on targeted and often transient protein interactions. We extensively investigate the biochemical processes of relevant human pathogens (Legionella pneumophila, Clostridioides difficile y Pseudomonas aeruginosa) y bacteria important to the environment (Dinoroseobacter shibae y Pseudomonas putida); (see posts: https://orcid.org/my-orcid?orcid=0000-0002-6409-8110). Dissecting the protein-protein interactions underlying such processes is crucial for basic research, but is also a pillar for multiple biomedical and biotechnological applications. We also investigate the molecular processes that govern the interactions of organisms in their natural niche. Our interest focuses on the elucidation of biogenesis pathways and cell-to-cell communication mechanisms driven by extracellular membrane vesicles (EMVs). VEMs are nanoparticles that are involved in a wide range of biological processes, such as horizontal DNA transfer, decoy function for phages and antibiotics, waste material removal and surface remodeling, nutrient removal, bacteria, immunomodulation of the host response and delivery of bioactive compounds, making them essential for communication between species and between kingdoms. Recently, our interactomics-based research has led us to the identification of highly conserved proteins that underpin the formation of VEMs across species. Currently, we propose to exploit the full potential of VEMs for the benefit of bacteria-plant interactions. To this end, we use interactomics techniques consisting of purification by affinity chromatography coupled to mass spectrometry, SPOT membrane matrices, immunofluorescence microscopy and immunolabeling with gold particles, among others, to identify other protein-protein interactions underlying the biogenesis of VEMs in different bacterial species present in the holobiont. Next, we induce the expression of these proteins found in the interactomic studies to modulate the rate, size and amount of vesicle formation. We intend to adapt the protein and metabolic load of VEMs by encapsulating nodulation factors, molecules for plant "vaccination", nitrogen-fixing enzymes or phytopathogen inhibitors. Thus, VEMs can be used as biopesticides to eliminate phytopathogens or as bioinoculants to improve nodulation, nitrogen fixation and, ultimately, plant growth. However, VEM research goes beyond applications related to sustainable agriculture, since the new molecular tools derived from this work can be applied in multiple disciplines, ranging from biomedicine (vaccine development) to bioremediation ( design of xenobiotic degraders based on VEMs).
The following techniques are available in our lab:
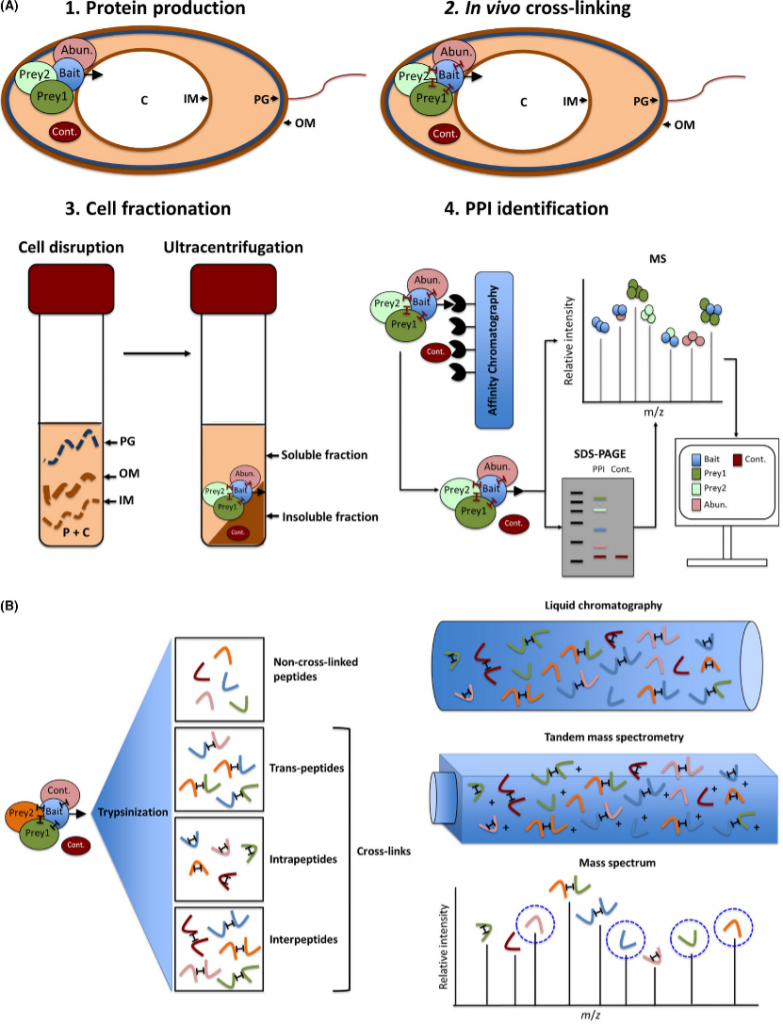
Figure 1. General interactomic workflow. The illustration depicts step by step the protein–protein interaction elucidation pathway via affinity chromatography copurification coupled with mass spectrometry (Borrero de Acuña, J. M. et al 2017).
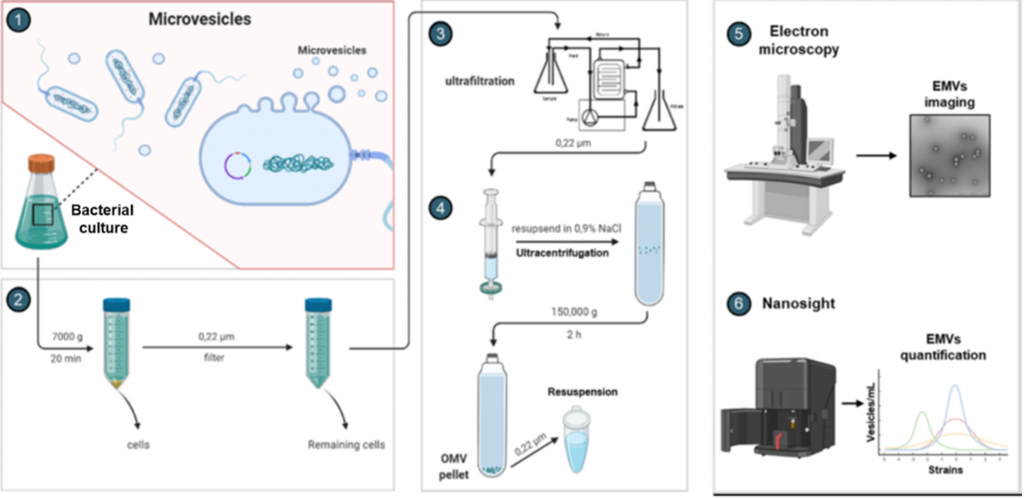
Figure 2. Experimental workflow designed for the isolation and characterisation of extracellular membrane vesicles (EMVs). (1) Growth of bacterial culture under standard conditions to produce EMVs. (2) Harvesting of cells and filtering of supernatants to remove remaining cells. (3) Ultrafiltration to remove impurities and concentrate EMV fractions. (4) Ultracentrifugation to collect EMV fractions. (5) Evaluation of the EMVs structure and integrity by electron microscopy. (6) Quantification of EMVs using scattering-light reliant Nanosight technology. Abbreviations: EMVs = extracellular membrane vesicles.
Research projects
- Title: Enhancing the symbiotic cross-talk through rhizobial membrane vesicles.
- Funding Entity: Ministry of Science and Innovation, Knowledge Generation Projects 2021, Government of Spain
- Reference: PID2021-122395OA-I00 Affiliation: Sevilla University Duration: September 2022-August 2026
- Funding Granted: 145.200,00€ Principal Investigator: Dr Jose Manuel Borrero de Acuña
- Title: Engineering membrane vesicles for fine-tuned modulation of rhizobia species interactions for enhanced nodulation and plant growth.
- Funding Entity: Junta de Andalucía, Ministry of economic transformation, industry, knowledge and universities
- Reference: EMERGIA20_00048 Affiliation: Sevilla University
- Duration: November 2021- August 2025 Principal Investigator: Dr Jose Manuel Borrero de Acuña
- Title: Developing a rodent disease model for Pseudomonas aeruginosa infection in bronchiectasis for drug research.
- Funding Entity: Federal Ministry of Education and Research (Germany)
- Reference: 281361126/GRK2223 Affiliation: Technical University Braunschweig
- Duration: December 2020- November 2023 Principal Investigator: Dr Jose Manuel Borrero de Acuña
Selected publications
- Jiménez-Guerrero, I., López-Baena, F.J., Borrero-de Acuña, J.M.* & Pérez-Montaño, F. (2023). Membrane vesicle engineering with «à la carte» bacterial-immunogenic molecules for
organism-free plant vaccination. Microbial Biotechnology, (accepted; ahead of print). *Corresponding author - Borrero‐from nowña, J. M., & Poblete‐Castro, I.* (2023). Rational engineering of natural polyhydroxyalkanoates producing microorganisms for improved synthesis and
recovery. Microbial Biotechnology, 16(2), 262-285. - Michel, A. M., Borrero‐from nowña, J. M.*, Molinari, G., … & Jahn, D. (15 authors); (2022).Cellular adaptation of Clostridioides difficile to high salinity encompasses a compatible solute‐
responsive change in cell morphology. Environmental Microbiology, 24(3), 1499-1517.*Corresponding author - Borrero‐from nowña, J. M., Gutierrez-Urrutia, I., Hidalgo-Dumont, C., ... & Poblete‐Castro, I.*; (14 authors); (2020). Channelling carbon flux through the meta‐cleavage route for improved
poly (3‐hydroxyalkanoate) production from benzoate and lignin‐based aromatics in Pseudomonas putida H. Microbial Biotechnology 14(6), 2385-2402. - Borrero-de Acuña, J.M., & Poblete-Castro, I.*; (2020). Expanding the Reach of Recombineering to Environmental Bacteria. Trends in Biotechnology, 38(7):684-685.
- Poblete-Castro, I., Aravena-Carrasco, C., Orellana-Saez, M., Pacheco, N., Cabrera, A., & Borrero-de Acuña, J.M.*; (2020). Engineering the osmotic state of Pseudomonas putida
KT2440 for efficient cell disruption and downstream processing of poly (3-hydroxyalkanoates). Frontiers in Bioengineering and Biotechnology, 8: 161. *Corresponding author - Borrero-de Acuña, J.M.*, Timmis, KN, Jahn, M. & Jahn, D. (2017). Protein complex formation during denitrification by Pseudomonas aeruginosa. Microbial Biotechnology, 10(6),
1523-1534. *Corresponding author - Borrero-de Acuña, J.M.*, Hidalgo-Dumont, C., Pacheco, N., Cabrera, A. & Poblete-Castro, I.* (2017). A novel programmable lysozyme-based lysis system in Pseudomonas
putida for biopolymer production. Scientific Reports, 29;7(1): 4373. *Co-Corresponding authors - Borrero-de Acuna, J.M., Rohde, M., Wissing, J., … & Jahn, D.*; (9 authors). (2016) Protein network of the Pseudomonas aeruginosa denitrification apparatus. Journal of
Bacteriology. 198(9): 1401-13
PhD students
- Irene Herrero Gómez: Enhancing the symbiotic cross-talk through rhizobial membrane vesicles.
- Natalia Moreno de Castro: Engineering membrane vesicles for fine-tuned modulation of rhizobia species interactions for enhanced nodulation and plant growth.
- Carlos Azogue Palma: Engineering membrane vesicles for efficient production of industrially valuable chemicals.
The type VI secretion system (T6SS) of Pseudomonas putida
IP: Patricia Bernal
Dr. Bernal's line of research focuses on the study of Type VI Secretion Systems (T6SS) in Pseudomonas putida. P. putida it is a soil bacterium with the ability to colonize the roots of different crop plants providing growth advantages and at the same time protection against pathogens; therefore this strain is considered an excellent agent of biological control (biocontrol). The biocontrol of diseases caused by plant pathogens is considered an excellent alternative to chemical pesticides to protect our crops, since these can cause contamination of the subsoil and the loss of the natural microbiota of both the soil and the plant. To progress in this field of research, it is key to understand in a global and complete way the molecular mechanisms of biological control carried out by known and well-established biocontrol agents such as Pseudomonas putida.
In P. putida, the T6SS has recently been established as a important biological control mechanism which gives this strain almost all of its biocontrolling capacity (Bernal et al. 2017). The T6SS is considered a powerful antibacterial weapon and P. putida uses it efficiently to kill extremely deleterious plant pathogens such as Pseudomonas syringae O Agrobacterium tumefaciens. P. putida It contains three T6SSs called K1-, K2-, and K3-T6SS. K1-T6SS is induced in a stationary growth phase, secretes toxins and kills a wide range of phytopathogenic bacteria such as P. syringae, Xanthomonas campestris O Agrobacterium tumefaciens (Bernal et al. 2017, Bernal et al. 2021). This antimicrobial capacity against phytopathogens has been observed both in experiments in vitro as in experiments in planta (Bernal et al. 2017, Bernal et al. 2021).
At a structural level, T6SS is a contractile nanomachine made up of a membrane component, a tail (contractile tube and sheath) and a base plate. The membrane component anchors the system to the cell envelope that brings together the base plate components from where the glue polymerizes. Once the system is assembled, the sheath contracts and the tube with the effector proteins is ejected into the target cells (Fig. 3). An elemental component of this system, the TssA protein, is essential for the assembly of the sheath. Despite their key function, TssA proteins exhibit unexpected diversity and exist in two main forms, a short form (TssAS) and a long form (TssAL). While the TssA proteinsL interact with the TagA protein to anchor the distal end of the extended sheath, the stabilization mechanism of T6SS containing TssAS it was still unknown. In a recent study (Bernal et al., 2021) we have identified a new category of structural components (TagB and TagJ proteins) that interact with TssA proteinss and contribute to the assembly of the T6SS by stabilizing the polymerized sheath from the base plate. In addition, we have shown that the presence of these components is important for full extension of the sheath and for maintaining optimal firing ability. Likewise, it has been seen that the association of different forms of TssA proteins with a different class of sheath stabilizing proteins results in T6SS that reside well in the cell for some time (TssAL-TagA) or fire immediately after extension of the sheath (TssAS-TagB). It is proposed that this diversity in shooting dynamics could contribute to the specialization of T6SS to adapt to different bacterial lifestyles in various environmental niches (Bernal et al., 2021).
The lines of research carried out in Dr. Bernal's laboratory are aimed at gaining a deeper understanding of the function and mechanism of action of these antimicrobial systems, which allows them to be adapted to develop highly effective biocontrol agents that are more specific. and with greater power to eradicate plant pathogens and be a tool for future biotechnological applications applied to agriculture.
In this video The project financed by the BBVA Foundation between 2021-2022 and titled: The biocontrol super agent Pseudomonas putida KT2440 as a sustainable measure to fight against Xylella fastidiosa, the great threat to our olive grove.
Active projects
- Title of the project: The T6SS antibacterial arsenal of the biocontrol agent Pseudomonas putida. Discovering novel compounds to fight antimicrobial resistance (AMR) and enhance crop protection.
- Financing entity: Ministry of Science and Innovation, Knowledge Generation Projects 2021, Government of Spain
- Reference: CNS2022-135585 Lead Entity: Sevilla University Period: September 2023-August 2025
- Subsidy amount: 199.154,00€ Responsible Researcher: Dr. Patricia Bernal
- Title of the project: Optimizing Pseudomonas putida as a crop protection agent: environmental signals, specificity and efficiency of the type vi secretion system as a biocontrol weapon
- Financing entity: Ministry of Science and Innovation, Knowledge Generation Projects 2021, Government of Spain
- Reference: PID2021-123000OB-I00 Lead Entity: Sevilla University Period: September 2022-August 2025
- Subsidy amount: 175.450,00€ Responsible Researcher: Dr. Patricia Bernal
- Title of the project: Biological control studies for crop protection through the use of the bacterial type VI secretion system (T6SS)
- Financing entity: Ministry of Science, Innovation and Universities, Ramón y Cajal Program Call 2019, Government of Spain
- Reference: RYC2019-026551-I Lead Entity: Sevilla University
- Period: April 2021-March 2026 Responsible Researcher: Dra. Patricia Bernal
Featured posts
- All mushrooms LP* & Bernal, P* (2023). Killing in the name of: T6SS structure and effectors diversity. Microbiology e1367. (*Corresponding co-author).
- Bernal, P*, Civantos, C, Pacheco-Sánchez, D, Quesada, JM, Filloux, A*, and Llamas*, MA (2023). Transcriptional organization and regulation of the Pseudomonas putida K1 Type VI secretion system gene cluster. Microbiology 169:001295. (*Co-author of correspondence).
- González-Magaña, A, Altuna, J, Queralt-Martín, M, Largo, E, Velázquez, C, Montánchez, I, Bernal, P, Alcaraz, A, Albesa-Jové, D (2022). The P. aeruginosa effector Tse5 forms ion-selective membrane pores that disrupt the membrane potential of intoxicated bacteria. Communications Biology. 5:1189.
- Duran, D, Bernal, P, David Vazquez-Arias, Blanco-Romero, E, Garrido-Sanz, D, Redondo-Nieto, M, Rivilla, R and, Martin, M. (2021). Pseudomonas fluorescens F113 type VI Secretion Systems mediate bacterial killing and adaption to the rhizosphere microbiome. Scientific Report 11:5772-5785.
- Bernal, P, Furniss, CD, Fecht, S, Leung, RCY, Spiga, L, Mavridou, DAI & Filloux. (2021). A novel stabilization mechanism for the type VI secretion system sheath. Proc Natl Acad Sci. 118:e2008500118
- Allsopp, LP, Bernal, P, Nolan, L & Filloux A. (2020). Causalities of War: The connection between T6SS & microbiota. Cell Microbiol 22:e13153.
- Bernal, P. *, Llamas, MA, Filloux, A. (2018). Type VI secretion Systems in plant-associated bacteria. Environ Microbiol 20: 1-15 (* Correspondence author).
- Bernal, P. *, Allsopp, L.P., Filloux, A., and Llamas M.A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME Journal 11: 972-987 (* Author of correspondence).
