
Group photo. Some members of the GFL group from left to right and from top to bottom: Oreto Antúnez, Daniel Medina, Fany Carrasco, Daniele Salmone, Toni Jordán, José García Martínez, Tianlu Li, Alvaro Castells, José E. Pérez Ortín, Ana Miguel, Paula Alepuz , Lucas Morales.
The Yeast Functional Genomics group (GFL, http://www.uv.es/gfl/), initially established by Dr. José E. Pérez Ortín in the Department of Biochemistry and Molecular Biology of the University of Valencia (UVEG ), began his research on yeast molecular biology in 1992. Starting in 1995, the work was oriented towards yeast genomics, participating in projects of genome sequencing and functional analysis of Saccharomyces cerevisiae. In 2000, Dr. José García Martínez joined; in 2004, Dr. Paula Alepuz and more recently Dr. Vicente Tordera and Vicente Arnau. Currently, the group also has 3 doctoral students and 1 technical research support staff.
Our group has chosen yeast as a model organism for the study of the control of gene expression S. cerevisiae, the best known eukaryote. The availability of genetic and genomic techniques and tools makes this organism the most flexible to carry out experiments on the functions of genes and proteins for practically all the fundamental processes of eukaryotic cells. Currently, we carry out the study of the control of gene expression in yeast through three main lines of research:
Development of genomic tools for the study of the turnover of mRNAs
Gene expression control is a multilevel process that affects all stages of transcription: from mRNA initiation to stability. The complexity of this phenomenon requires, to be understood, new approaches, including those at the genomic scale. This approach can only be done in a model organism that has the necessary tools, such as yeast. S. cerevisiae. In our group we develop genomic tools in yeast to study transcription in vivo and clarify the functional networks that control genes and transcription on a global scale. The objective of this line of research is to define the mechanisms that regulate gene expression during transcriptional elongation and the relationship between transcription control mechanisms and the degradation of messengers for the homeostatic control of their levels. We are also adapting these study tools in the model pathogenic yeast Candida albicans.
Coordination mechanisms between transcription and mRNA stability in yeast during the stress response
Responses to stress are complex and span many levels. One of the most relevant is that of the transcriptional response. With the genomic tools developed in our laboratory, we address the relative importance of the synthesis and degradation of mRNAs in different stress situations of practical importance in biomedicine or biotechnology: the response to osmotic stress, the response of wine strains of S. cerevisiae to nitrogen deficiency in musts or the response of C. albicans to oxidative stress caused by the host.
Molecular mechanisms involved in the post-transcriptional regulation of gene expression in response to environmental signals: regulation of splicing, stability and translation of messengers in response to osmotic stress in yeast S. cerevisiae
In recent years, it has been shown that post-transcriptional regulation occurs in parallel to transcriptional regulation in response to many extracellular signals and, in many cases, especially in mammals, post-transcriptional mechanisms are the ones that have a greater weight. in adaptive responses. Thus, for the regulation of the amount of mRNA in the cell, the variation of the splicing efficiency of the pre-mRNA and the degradation rate of the mature mRNAs are used, which, added to the regulation of their translation rates, allows quickly modify protein levels in the cell to generate an adequate response to each stimulus. The objective of this line of research is to discover the mechanisms involved in the post-translational regulation of mRNAs of S. cerevisiae in response to osmotic stress.
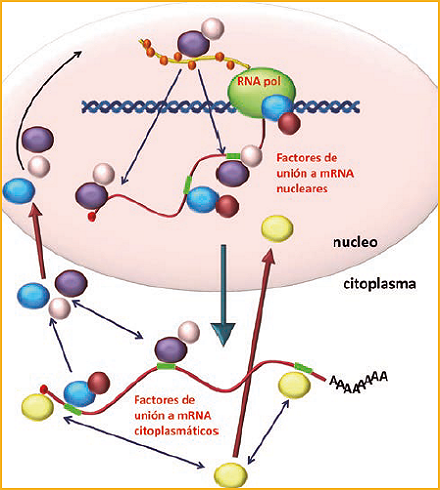
Figure 1. Model of exchange of mRNA binding factors during the different stages of the gene expression process in eukaryotes.
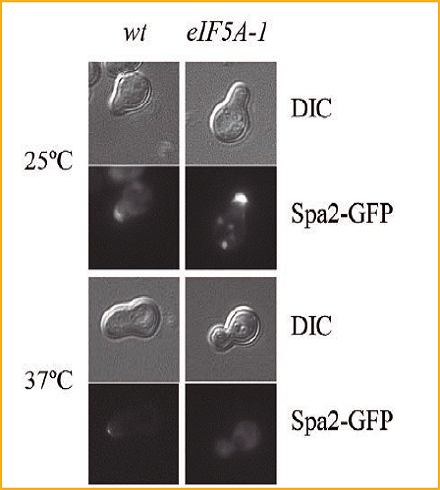
Figure 2. The elongation factor eIF5A, conserved from yeast to humans, is necessary for shmoos formation and polarized growth in the pheromone response of S. cerevisiae cells. The photo shows how the localization at the tip of the shmoo of a polarisome protein, Spa2, fused to GFP, which is produced after pheromone treatment, is lost at restrictive temperature in a temperature-sensitive mutant of eIF5A.
Throughout these years, we have established collaborations with researchers from national and international groups such as:
- Dr. Joaquín Moreno, Department of Biochemistry and Molecular Biology (UVEG).
- Dr. Per Sunnerhagen and Dr. Jonas Warringer. (Gothenburg University, Sweden).
- Dr. Gustav Ammerer (University of Vienna, Austria).
- Dr. Sebastián Chávez (University of Seville).
- Dr. Francisco Navarro (University of Jaén).
- Dr. Enrique Herrero (University of Lleida).
- Dr. Jesús Pla (Complutense University of Madrid).
- Dr. Joachim Ernst (Heinrich Heine University Düsseldorf)
- Dr. Joaquín Ariño (Autonomous University of Barcelona)
- Dr. Mordechai Choder (Technion, Israel)
- Dra. Alexandra Mendes-Ferrerira (U. Tras-os-Montes and Alto Douro, Portugal)
In the last five years we have received funding through four projects of the National R + D + i Plan, ten research grants from the Generalitat Valenciana and a subsidy from the UVEG. The GFL group is a member of ISIC / ERI BIOTECMED, Higher Institute for Cooperative Research of the Generalitat Valenciana and Interdisciplinary Research Structure of the UVEG in Biotechnology and Biomedicine (http://www.uv.es/biotecmed). It is also part of the Biotechnology and Biomedicine Microcluster with Model Yeasts (BBLM) made up of groups from the UVEG, the Polytechnic University of Valencia, the CSIC and the Príncipe Felipe Research Center, within the Campus of International Excellence «VLC Campus» (http://www.vlc-campus.com/).
Representative bibliography
Magraner-Pardo L, Pelechano V, Coloma MD, Tordera V. (2014). Dynamic remodeling of histone modifications in response to osmotic stress in Saccharomyces cerevisiae. BMC Genomics.15:247.
Li T, Belda-Palazón B, Ferrando A, Alepuz P. (2014). Fertility and Polarized Cell Growth Depends on eIF5A for Translation of Polyproline-Rich Formins in Saccharomyces cerevisiae. Genetics. 197:1191-1200.
Miguel A, Montón F, Li T, Gómez-Herreros F, Chávez S, Alepuz P and Pérez-Ortín JE. (2013). External conditions inversely change the RNA polymerase II elongation rate and density in yeast. Biochim Biophys Acta 11:1248-55.
Nikolaou C, Bermúdez I, Manichanh C, García-Martinez J, Guigó R, Pérez- Ortín JE and Roca J. Remuzgo-Martínez S, Pilares-Ortega L, Icardo JM, Valdizán EM, Vargas VI, Pazos A, Ramos-Vivas J.
Topoisomerase II regulates yeast genes with singular chromatin architectures. Nucl Acids Res 41: 9243-9256.
Pérez-Ortín JE, Alepuz P, Chávez S and Choder M. (2013). Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J Mol Biol. 425:3750-3775
Pérez-Ortín JE, Medina DA, Chávez S and Moreno J. (2013). What do you mean by transcription rate?: The conceptual difference between nascent transcription rate and mRNA synthesis rate is essential for the proper understanding of transcriptomic analyses. Bioessays 35:1056-1062.
Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X and Choder M. (2013). Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153:1000-1011.
Garre E, Romero-Santacreu L, Barneo-Muñoz M, Miguel A, Pérez-Ortín JE and Alepuz P. (2013). Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress. PLoS ONE DOI: 10.1371/journal.pone.0061240.
Alic A, Pérez-Ortín JE, Moreno J and Arnau V. (2013). mRNAStab–a web application for mRNA stability analysis. Bioinformatics 29:813-814.
García-Martínez, J, Ayala, G, Pelechano, V, Chávez, S, Herrero, E, and Pérez-Ortín, JE. (2012). The relative importance of transcription rate, cryptic transcription and mRNA stability on shaping stress responses in yeast. Transcription 3: 39-44.
Pérez-Ortín, JE, De Miguel-Jiménez y Chávez, S (2012). Genome-wide studies of mRNA synthesis and degradation in eukaryotes. Biochim. Biophys. Acta 1819: 604-615.
Castells-Roca L, García-Martínez J, Moreno J, Herrero E, Bellí, G and Pérez-Ortín, JE. (2011). Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLoS ONE 6: e17272.
García-López, MC, Pelechano V, Mirón-García MC. Garrido-Godino AI, Werner M, Pérez-Ortín JE and Navarro F. (2011). The Conserved Foot Domain of RNA Pol II Associates with Proteins Involved in Transcriptional Initiation and/or Early Elongation. Genetics 189: 1235-1248.
García-Martínez J, Ayala G, Pelechano V, Chávez S, Herrero E and Pérez-Ortín JE. (2012). The relative importance of transcription rate, cryptic transcription and mRNA stability on shaping stress responses in yeast. Transcription 3: 39-44.
Pérez-Ortín JE., By Miguel-Jiménez and Chávez S. (2012). Genome-wide studies of mRNA synthesis and degradation in eukaryotes. Biochim. Biophys. Acta 1819: 604-615.
Rodríguez-Gil A, García-Martínez J, Pelechano V, Muñoz-Centeno M.C, Geli V, Pérez-Ortín JE. and Chávez S. (2010). The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors. Nucleic Acids Res. 38:4651-64
Pelechano V, Chávez S. and Pérez-Ortín JE. (2010). A complete set of nascent transcription rates for yeast genes. PLoS ONE 5: e15442.
Romero-Santacreu L, Orozco H, Garre E and Alepuz P. (2010). The bidirectional cytomegalovirus immediate/early promoter is regulated by Hog1 and the stress transcription factors Sko1 and Hot1 in yeast. Mol. Gen. Genom. 283: 510-518.
Romero-Santacreu L, Moreno J, Pérez-Ortín, JE and Alepuz P. (2009). Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15:1110–1120.
Pelechano V, Jimeno-González S, Rodríguez-Gil A, García-Martínez J, Pérez-Ortín, JE and Chávez S. (2009). Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 5(8):e1000614.
